6ICZ
 
 | | Cryo-EM structure of a human post-catalytic spliceosome (P complex) at 3.0 angstrom | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
5C7G
 
 | |
6ZTQ
 
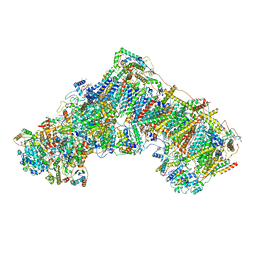 | | Cryo-EM structure of respiratory complex I from Mus musculus inhibited by piericidin A at 3.0 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Bridges, H.R, Blaza, J.N, Agip, A.N.A, Hirst, J. | | Deposit date: | 2020-07-20 | | Release date: | 2020-10-21 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of inhibitor-bound mammalian complex I.
Nat Commun, 11, 2020
|
|
5C87
 
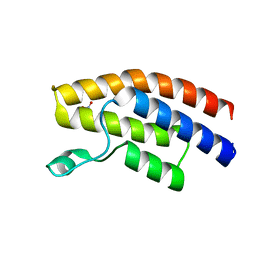 | |
6THY
 
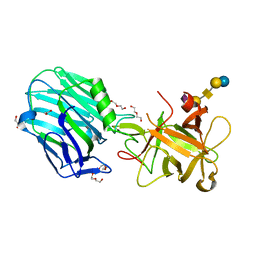 | | Botulinum neurotoxin A3 Hc domain in complex with GD1a | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BoNT/A3, ... | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M. | | Deposit date: | 2019-11-21 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of botulinum neurotoxin subtype A3 cell binding domain in complex with GD1a co-receptor ganglioside.
Febs Open Bio, 10, 2020
|
|
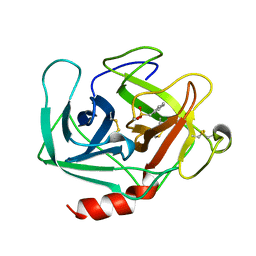
1KLT
 
 | |
6EEV
 
 | | Structure of class II HMG-CoA reductase from Delftia acidovorans with mevalonate bound | | Descriptor: | (R)-MEVALONATE, 3-hydroxy-3-methylglutaryl coenzyme A reductase, GLYCEROL, ... | | Authors: | Ragwan, E.R, Arai, E, Kung, Y. | | Deposit date: | 2018-08-15 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | New Crystallographic Snapshots of Large Domain Movements in Bacterial 3-Hydroxy-3-methylglutaryl Coenzyme A Reductase.
Biochemistry, 57, 2018
|
|
4ZAX
 
 | | Structure of UbiX in complex with oxidised prenylated FMN (radical) | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, PHOSPHATE ION, THIOCYANATE ION, ... | | Authors: | White, M.D, Leys, D. | | Deposit date: | 2015-04-14 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
Nature, 522, 2015
|
|
1KTX
 
 | | KALIOTOXIN (1-37) SHOWS STRUCTURAL DIFFERENCES WITH RELATED POTASSIUM CHANNEL BLOCKERS | | Descriptor: | KALIOTOXIN | | Authors: | Fernandez, I, Romi, R, Szendefi, S, Martin-Eauclaire, M.-F, Rochat, H, Van Rietschtoten, J, Pons, M, Giralt, E. | | Deposit date: | 1994-06-02 | | Release date: | 1995-01-26 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Kaliotoxin (1-37) shows structural differences with related potassium channel blockers.
Biochemistry, 33, 1994
|
|
8I5Y
 
 | | Structure of human Nav1.7 in complex with vixotrigine | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-01-26 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
4M8Y
 
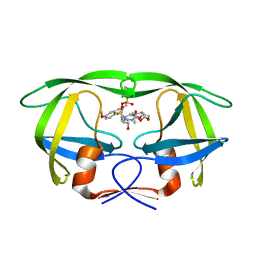 | | GS-8374, a Novel Phosphonate-Containing Inhibitor of HIV-1 Protease, Effectively Inhibits HIV PR Mutants with Amino Acid Insertions | | Descriptor: | DIETHYL ({4-[(2S,3R)-2-({[(3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YLOXY]CARBONYL}AMINO)-3-HYDROXY-4-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}BUTYL]PHENOXY}METHYL)PHOSPHONATE, Protease | | Authors: | Grantz saskova, K, Brynda, J, Rezacova, P, Kozisek, M, Konvalinka, J. | | Deposit date: | 2013-08-14 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | GS-8374, a prototype phosphonate-containing inhibitor of HIV-1 protease, effectively inhibits protease mutants with amino acid insertions.
J.Virol., 88, 2014
|
|
1KRN
 
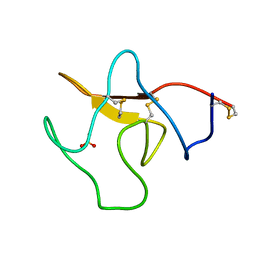 | | STRUCTURE OF KRINGLE 4 AT 4C TEMPERATURE AND 1.67 ANGSTROMS RESOLUTION | | Descriptor: | PLASMINOGEN, SULFATE ION | | Authors: | Stec, B, Teeter, M.M, Whitlow, M, Yamano, A. | | Deposit date: | 1995-06-21 | | Release date: | 1997-01-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure of human plasminogen kringle 4 at 1.68 a and 277 K. A possible structural role of disordered residues.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1KVX
 
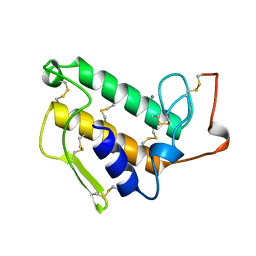 | |
5ETY
 
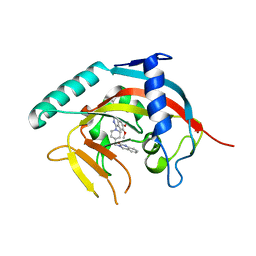 | | Crystal Structure of human Tankyrase-1 bound to K-756 | | Descriptor: | 3-[[1-(6,7-dimethoxyquinazolin-4-yl)piperidin-4-yl]methyl]-1,4-dihydroquinazolin-2-one, Tankyrase-1, ZINC ION | | Authors: | Takahashi, Y, Miyagi, H, Suzuki, M, Saito, J. | | Deposit date: | 2015-11-18 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Discovery and Characterization of K-756, a Novel Wnt/ beta-Catenin Pathway Inhibitor Targeting Tankyrase
Mol.Cancer Ther., 15, 2016
|
|
5T6Y
 
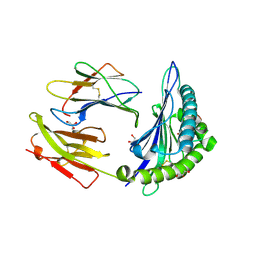 | | HLA-B*57:01 presenting TSTFEDVKILAF | | Descriptor: | ACETATE ION, Beta-2-microglobulin, Decapeptide: THR-SER-THR-PHE-GLU-ASP-VAL-LYS-ILE-LEU-ALA-PHE, ... | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2016-09-02 | | Release date: | 2017-03-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape.
Nat. Struct. Mol. Biol., 24, 2017
|
|
1KKH
 
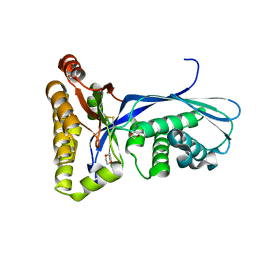 | | Crystal Structure of the Methanococcus jannaschii Mevalonate Kinase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Mevalonate Kinase | | Authors: | Yang, D, Shipman, L.W, Roessner, C.A, Scott, A.I, Sacchettini, J.C. | | Deposit date: | 2001-12-08 | | Release date: | 2002-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Methanococcus jannaschii mevalonate kinase, a member of the GHMP kinase superfamily.
J.Biol.Chem., 277, 2002
|
|
4MHZ
 
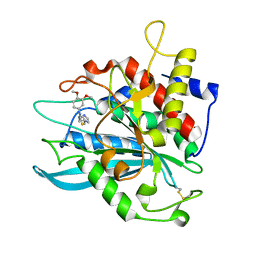 | | Crystal structure of apo-form glutaminyl cyclase from Ixodes scapularis in complex with PBD150 | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, Glutaminyl cyclase, putative | | Authors: | Huang, K.F, Hsu, H.L, Wang, A.H.J. | | Deposit date: | 2013-08-30 | | Release date: | 2014-03-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and functional analyses of a glutaminyl cyclase from Ixodes scapularis reveal metal-independent catalysis and inhibitor binding.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5EU9
 
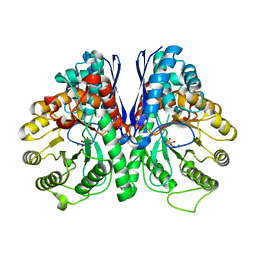 | | Structure of Human Enolase 2 in complex with ((3S,5S)-1,5-dihydroxy-3-methyl-2-oxopyrrolidin-3-yl)phosphonic acid | | Descriptor: | ((3S,5S)-1,5-dihydroxy-3-methyl-2-oxopyrrolidin-3-yl)phosphonic acid, Gamma-enolase, MAGNESIUM ION, ... | | Authors: | Leonard, P.G, Muller, F.L. | | Deposit date: | 2015-11-18 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | SF2312, a natural phosphonate inhibitor of Enolase
To be Published
|
|
5TAM
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 4) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5ESS
 
 | | Crystal Structure of M. tuberculosis MenD bound to Mg2+ and covalent intermediate I (a ThDP and decarboxylated 2-oxoglutarate adduct) | | Descriptor: | 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, 4-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-1,3-thiazol-3 -ium-2-yl]-4-oxidanyl-butanoic acid, ACETATE ION, ... | | Authors: | Johnston, J.M, Jirgis, E.N.M, Bashiri, G, Bulloch, E.M.M, Baker, E.N. | | Deposit date: | 2015-11-17 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Views along the Mycobacterium tuberculosis MenD Reaction Pathway Illuminate Key Aspects of Thiamin Diphosphate-Dependent Enzyme Mechanisms.
Structure, 24, 2016
|
|
6SHN
 
 | | Escherichia coli AGPase in complex with FBP. Symmetry C1 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Glucose-1-phosphate adenylyltransferase | | Authors: | Cifuente, J.O, Comino, N, D'Angelo, C, Marina, A, Gil-Carton, D, Albesa-Jove, D, Guerin, M.E. | | Deposit date: | 2019-08-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM.
Curr Res Struct Biol, 2, 2020
|
|
3ZU8
 
 | | STRUCTURE OF CBM3B OF MAJOR SCAFFOLDIN SUBUNIT SCAA FROM ACETIVIBRIO CELLULOLYTICUS DETERMINED ON THE NIKEL ABSORPTION EDGE | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CELLULOSOMAL SCAFFOLDIN, ... | | Authors: | Yaniv, O, Halfon, Y, Lamed, R, Frolow, F. | | Deposit date: | 2011-07-17 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of Cbm3B of the Major Scaffoldin Subunit Scaa from Acetivibrio Cellulolyticus
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4LP6
 
 | | Crystal Structure of Human Carbonic Anhydrase II in complex with a quinoline oligoamide foldamer | | Descriptor: | 8-({[4-(3-aminopropoxy)-8-({[4-hydroxy-8-({[4-(2-methylpropoxy)-8-({[4-(3-{[(4-sulfamoylbenzoyl)amino]methyl}phenoxy)butyl]carbamoyl}amino)quinolin-2-yl]carbonyl}amino)quinolin-2-yl]carbonyl}amino)quinolin-2-yl]carbonyl}amino)-4-(carboxymethoxy)quinoline-2-carboxylic acid, Carbonic anhydrase 2, ZINC ION | | Authors: | Buratto, J, Granier, T, Langlois D'estaintot, B, Huc, I, Gallois, B. | | Deposit date: | 2013-07-15 | | Release date: | 2013-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of a complex formed by a protein and a helical aromatic oligoamide foldamer at 2.1 angstrom resolution.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
8I0T
 
 | | The cryo-EM structure of human Bact-III complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Crooked neck-like protein 1, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8BE8
 
 | | Crystal structure of SOS1-HRas-peptidomimetic4 | | Descriptor: | FORMIC ACID, GTPase HRas, SOS1-HRas-peptidomimetic4, ... | | Authors: | Fischer, B, Wohlkonig, A, Steyaert, J. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nanobody Loop Mimetics Enhance Son of Sevenless 1-Catalyzed Nucleotide Exchange on RAS.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
