4CH4
 
 | | Structure of pyrrolysyl-tRNA synthetase in complex with adenylated crotonyl lysine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[({(2R)-2-amino-6-[(2E)-but-2-enoylamino]hexanoyl}oxy)phosphinato]adenosine, MAGNESIUM ION, ... | | Authors: | Fluegel, V, Vrabel, M, Schneider, S. | | Deposit date: | 2013-11-28 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Basis for the Site-Specific Incorporation of Lysine Derivatives Into Proteins.
Plos One, 9, 2014
|
|
1MCT
 
 | | THE REFINED 1.6 ANGSTROMS RESOLUTION CRYSTAL STRUCTURE OF THE COMPLEX FORMED BETWEEN PORCINE BETA-TRYPSIN AND MCTI-A, A TRYPSIN INHIBITOR OF SQUASH FAMILY | | Descriptor: | BETA-TRYPSIN, CALCIUM ION, TRYPSIN INHIBITOR A | | Authors: | Huang, Q, Liu, S, Tang, Y. | | Deposit date: | 1992-10-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Refined 1.6 A resolution crystal structure of the complex formed between porcine beta-trypsin and MCTI-A, a trypsin inhibitor of the squash family. Detailed comparison with bovine beta-trypsin and its complex.
J.Mol.Biol., 229, 1993
|
|
6UME
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | 2-(pyridin-3-yl)-N-(5-{4-[(5-{[(pyridin-3-yl)acetyl]amino}-1,3,4-thiadiazol-2-yl)amino]piperidin-1-yl}-1,3,4-thiadiazol-2-yl)acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-09 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00012
To Be Published
|
|
6BD7
 
 | | Crystal structure of human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-{[(2R)-3-oxo-2-[(propan-2-yl)amino]-3-{[(pyridin-3-yl)methyl]amino}propyl]sulfanyl}-3-phenylpropan-2-yl]carbamate | | Authors: | Sevrioukova, I. | | Deposit date: | 2017-10-21 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Interaction of the rationally designed ritonavir-like inhibitors with human cytochrome P450 3A4: Impact of the side group interplay
Mol. Pharm., 2017
|
|
4NE2
 
 | | Pantothenamide-bound Pantothenate Kinase from Klebsiella pneumoniae | | Descriptor: | (R)-N-(3-((2-(benzo[d][1,3]dioxol-5-yl)ethyl)amino)-3-oxopropyl)-2,4-dihydroxy-3,3-dimethylbutanamide, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hughes, S.J, Antoshchenko, T, Kim, K.P, Smil, D, Park, H.W. | | Deposit date: | 2013-10-28 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of a new N-substituted pantothenamide bound to pantothenate kinases from Klebsiella pneumoniae and Staphylococcus aureus.
Proteins, 82, 2014
|
|
5NUU
 
 | | Torpedo californica acetylcholinesterase in complex with a chlorotacrine-tryptophan hybrid inhibitor | | Descriptor: | (2~{S})-2-azanyl-~{N}-[6-[(6-chloranyl-1,2,3,4-tetrahydroacridin-9-yl)amino]hexyl]-3-(1~{H}-indol-3-yl)propanamide, Acetylcholinesterase, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Caliandro, R, Pesaresi, A, Lamba, D. | | Deposit date: | 2017-05-02 | | Release date: | 2018-05-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel tacrine-tryptophan hybrids: Multi-target directed ligands as potential treatment for Alzheimer's disease.
Eur.J.Med.Chem., 168, 2019
|
|
3M32
 
 | | Structural Insight into Methyl-Coenzyme M Reductase Chemistry using Coenzyme B Analogues | | Descriptor: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Cedervall, P.E, Dey, M, Ragsdale, S.W, Wilmot, C.M. | | Deposit date: | 2010-03-08 | | Release date: | 2010-09-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insight into methyl-coenzyme M reductase chemistry using coenzyme B analogues.
Biochemistry, 49, 2010
|
|
5EQE
 
 | | Crystal structure of choline kinase alpha-1 bound by [4-[(4-methyl-1,4-diazepan-1-yl)methyl]phenyl]methanamine (compound 11) | | Descriptor: | Choline kinase alpha, [4-[(4-methyl-1,4-diazepan-1-yl)methyl]phenyl]methanamine | | Authors: | Zhou, T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel Small Molecule Inhibitors of Choline Kinase Identified by Fragment-Based Drug Discovery.
J.Med.Chem., 59, 2016
|
|
7XGG
 
 | |
4BGH
 
 | | Crystal Structure of CDK2 in complex with pan-CDK Inhibitor | | Descriptor: | 4-((5-BROMO-4-(PROP-2-YN-1-YLAMINO)PYRIMIDIN-2-YL)AMINO)BENZENESULFONAMIDE, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Luecking, U, Jautelat, R, Krueger, M, Brumby, T, Lienau, P, Schaefer, M, Briem, H, Schulze, J, Hillisch, A, Reichel, A, Siemeister, G. | | Deposit date: | 2013-03-26 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Lab Oddity Prevails: Discovery of Pan-Cdk Inhibitor (R)- S-Cyclopropyl-S-(4-{[4-{[(1R,2R)-2-Hydroxy-1-Methylpropyl]Oxy}-5-(Trifluoromethyl)Pyrimidin-2-Yl]Amino}Phenyl)Sulfoximide (Bay 1000394) for the Treatment of Cancer.
Chemmedchem, 8, 2013
|
|
5UKF
 
 | | Crystal Structure of the Human Vaccinia-related Kinase 1 Bound to an Oxindole Inhibitor | | Descriptor: | 4-{[(Z)-(7-oxo-6,7-dihydro-8H-[1,3]thiazolo[5,4-e]indol-8-ylidene)methyl]amino}benzene-1-sulfonamide, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Counago, R.M, Wells, C, Zuercher, W, Willson, T.M, Bountra, C, Edwards, A.M, Arruda, P, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-22 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of human Vaccinia-Related Kinases (VRK) bound to small-molecule inhibitors identifies different P-loop conformations.
Sci Rep, 7, 2017
|
|
9FY5
 
 | | CRYSTAL STRUCTURE OF HUMAN MONOACYLGLYCEROL LIPASE WITH COMPOUND | | Descriptor: | (4aR,8aS)-6-[4-(diphenylmethyl)piperidin-1-yl]carbonyl-4,4a,5,7,8,8a-hexahydropyrido[4,3-b][1,4]oxazin-3-one, 1,2-ETHANEDIOL, Monoglyceride lipase | | Authors: | Gazzi, T, Grether, U, Nazare, M, Hochstrasser, R, Wang, H, Heer, D, Topp, A, Benz, J. | | Deposit date: | 2024-07-02 | | Release date: | 2024-12-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Development of a Highly Selective NanoBRET Probe to Assess MAGL Inhibition in Live Cells.
Chembiochem, 26, 2025
|
|
5ELJ
 
 | | Isoform-specific inhibition of SUMO-dependent protein-protein interactions | | Descriptor: | SUMO-Affirmer-S2D5, Small ubiquitin-related modifier 1 | | Authors: | Hughes, D.J, Tiede, C, Hall, N, Tang, A.A.S, Trinh, C.H, Zajac, K, Mandal, U, Howell, G, Edwards, T.A, McPherson, M.J, Tomlinson, D.C, Whitehouse, A. | | Deposit date: | 2015-11-04 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Generation of specific inhibitors of SUMO-1- and SUMO-2/3-mediated protein-protein interactions using Affimer (Adhiron) technology.
Sci Signal, 10, 2017
|
|
3K93
 
 | |
9FFJ
 
 | | Artificial metalloenzyme with a nickel-based 1,10-phenanthroline cofactor and streptavidin N49M-S112V mutant | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lau, K, Wang, W, Pojer, F, Larabi, A. | | Deposit date: | 2024-05-23 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Artificial Metalloenzymes with Two Catalytic Cofactors for Tandem Abiotic Transformations.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
4N7Q
 
 | | Crystal structure of eukaryotic THIC from A. thaliana | | Descriptor: | COBALT (II) ION, HEXANE-1,6-DIOL, Phosphomethylpyrimidine synthase, ... | | Authors: | Coquille, S.C, Roux, C, Mehta, A, Begley, T.P, Fitzpatrick, T.B, Thore, S. | | Deposit date: | 2013-10-16 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution crystal structure of the eukaryotic HMP-P synthase (THIC) from Arabidopsis thaliana.
J.Struct.Biol., 184, 2013
|
|
2VWY
 
 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION, N'-(5-chloro-1,3-benzodioxol-4-yl)-N-(3-methylsulfonylphenyl)pyrimidine-2,4-diamine | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Leach, A.G, Kettle, J.G. | | Deposit date: | 2008-06-30 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 2: Structure-Based Discovery and Optimisation of 3,5-Bis Substituted Anilinopyrimidines.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5CSF
 
 | | S100B-RSK1 crystal structure A | | Descriptor: | CALCIUM ION, Protein S100-B, Ribosomal protein S6 kinase alpha-1 | | Authors: | Gogl, G, Nyitray, L. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Ribosomal S6 Kinase 1 (RSK1) Inhibition by S100B Protein: MODULATION OF THE EXTRACELLULAR SIGNAL-REGULATED KINASE (ERK) SIGNALING CASCADE IN A CALCIUM-DEPENDENT WAY.
J.Biol.Chem., 291, 2016
|
|
7XJ8
 
 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 2 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 2 conformation
To Be Published
|
|
5CXB
 
 | |
3JRR
 
 | | Crystal structure of the ligand binding suppressor domain of type 3 inositol 1,4,5-trisphosphate receptor | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 3 | | Authors: | Chan, J, Ishiyama, N, Ikura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A 1.9 angstrom crystal structure of the suppressor domain of type 3 inositol 1,4,5-trisphosphate receptor
To be Published
|
|
5B2R
 
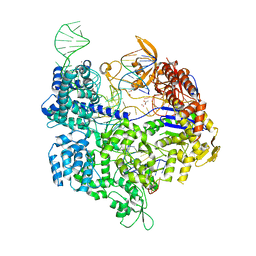 | | Crystal structure of the Streptococcus pyogenes Cas9 VQR variant in complex with sgRNA and target DNA (TGA PAM) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CRISPR-associated endonuclease Cas9, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Altered PAM Specificities of Engineered CRISPR-Cas9
Mol.Cell, 61, 2016
|
|
1LF7
 
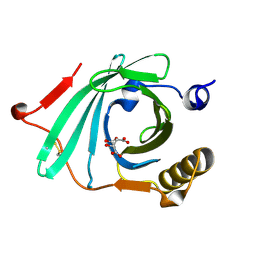 | | Crystal Structure of Human Complement Protein C8gamma at 1.2 A Resolution | | Descriptor: | CITRIC ACID, Complement Protein C8gamma | | Authors: | Ortlund, E, Parker, C.L, Schreck, S.F, Ginell, S, Minor, W, Sodetz, J.M, Lebioda, L. | | Deposit date: | 2002-04-10 | | Release date: | 2002-06-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of human complement protein C8gamma at 1.2 A resolution reveals a lipocalin fold and a distinct ligand binding site.
Biochemistry, 41, 2002
|
|
7KNO
 
 | |
7KNP
 
 | |
