4UZG
 
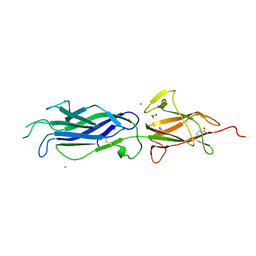 | | Crystal structure of group B streptococcus pilus 2b backbone protein SAK_1440 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SURFACE PROTEIN SPB1 | | Authors: | Malito, E, Cozzi, R, Bottomley, M.J. | | Deposit date: | 2014-09-05 | | Release date: | 2015-05-13 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Structure and assembly of group B streptococcus pilus 2b backbone protein.
PLoS ONE, 10, 2015
|
|
7CJD
 
 | |
9FQJ
 
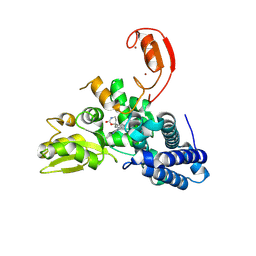 | | E3 ligase Cbl-b in complex with a carbamate scaffold inhibitor (compound 12) | | Descriptor: | 2-cyclopropyl-6-methyl-~{N}-[3-[(6~{S})-6-methyl-2-oxidanylidene-1,3-oxazinan-6-yl]phenyl]pyrimidine-4-carboxamide, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2024-06-17 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.563 Å) | | Cite: | Accelerated Discovery of a Carbamate Scaffold Cbl-b Inhibitor using Generative Models and Structure-Based Drug Design
To be published
|
|
7CW3
 
 | | Cryo-EM structure of Chikungunya virus in complex with mAb CHK-263 IgG (subregion around icosahedral 2-fold vertex) | | Descriptor: | Capsid protein, E1 glycoprotein, E2 glycoprotein, ... | | Authors: | Zhou, Q.F, Fox, J.M, Earnest, J.T, Ng, T.S, Kim, A.S, Fibriansah, G, Kostyuchenko, V.A, Shu, B, Diamond, M.S, Lok, S.M. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structural basis of Chikungunya virus inhibition by monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7CMD
 
 | | Crystal structure of the SARS-CoV-2 PLpro with GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, ZINC ION | | Authors: | Gao, X, Cui, S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of SARS-CoV-2 papain-like protease.
Acta Pharm Sin B, 11, 2021
|
|
2B7T
 
 | | Structure of ADAR2 dsRBM1 | | Descriptor: | Double-stranded RNA-specific editase 1 | | Authors: | Stefl, R, Xu, M, Skrisovska, L, Emeson, R.B, Allain, F.H.-T. | | Deposit date: | 2005-10-05 | | Release date: | 2006-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and specific RNA binding of ADAR2 double-stranded RNA binding motifs.
Structure, 14, 2006
|
|
7CW2
 
 | | Cryo-EM structure of Chikungunya virus in complex with Fab fragments of mAb CHK-263 (subregion around icosahedral 5-fold vertex) | | Descriptor: | Capsid protein, E1 glycoprotein, E2 glycoprotein, ... | | Authors: | Zhou, Q.F, Fox, J.M, Earnest, J.T, Ng, T.S, Kim, A.S, Fibriansah, G, Kostyuchenko, V.A, Shu, B, Diamond, M.S, Lok, S.M. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of Chikungunya virus inhibition by monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2B7V
 
 | | Structure of ADAR2 dsRBM2 | | Descriptor: | Double-stranded RNA-specific editase 1 | | Authors: | Stefl, R, Xu, M, Skrisovska, L, Emeson, R.B, Allain, F.H.-T. | | Deposit date: | 2005-10-05 | | Release date: | 2006-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and specific RNA binding of ADAR2 double-stranded RNA binding motifs.
Structure, 14, 2006
|
|
9FQI
 
 | | E3 ligase Cbl-b in complex with a lactam scaffold inhibitor (compound 7) | | Descriptor: | 8-[3-[(4~{R})-4-methyl-2-oxidanylidene-piperidin-4-yl]phenyl]-3-[[(3~{S})-3-methylpiperidin-1-yl]methyl]-5-(trifluoromethyl)-1$l^{4},7,8-triazabicyclo[4.3.0]nona-1(6),2,4-trien-9-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2024-06-17 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Accelerated Discovery of a Carbamate Scaffold Cbl-b Inhibitor using Generative Models and Structure-Based Drug Design
To be published
|
|
9FQH
 
 | | E3 ligase Cbl-b in complex with a triazolone core inhibitor (compound 1) | | Descriptor: | 8-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-3-[[(3~{S})-3-methylpiperidin-1-yl]methyl]-5-(trifluoromethyl)-1$l^{4},7,8-triazabicyclo[4.3.0]nona-1(6),2,4-trien-9-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2024-06-17 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Accelerated Discovery of a Carbamate Scaffold Cbl-b Inhibitor using Generative Models and Structure-Based Drug Design
To be published
|
|
9F7X
 
 | |
7CW0
 
 | | Cryo-EM structure of Chikungunya virus in complex with mAb CHK-263 IgG | | Descriptor: | Capsid protein, E1 glycoprotein, E2 glycoprotein, ... | | Authors: | Zhou, Q.F, Fox, J.M, Earnest, J.T, Ng, T.S, Kim, A.S, Fibriansah, G, Kostyuchenko, V.A, Shu, B, Diamond, M.S, Lok, S.M. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of Chikungunya virus inhibition by monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7CVZ
 
 | | Cryo-EM structure of Chikungunya virus in complex with Fab fragments of mAb CHK-263 | | Descriptor: | Capsid protein, E1 glycoprotein, E2 glycoprotein, ... | | Authors: | Zhou, Q.F, Fox, J.M, Earnest, J.T, Ng, T.S, Kim, A.S, Fibriansah, G, Kostyuchenko, V.A, Shu, B, Diamond, M.S, Lok, S.M. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis of Chikungunya virus inhibition by monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7CVY
 
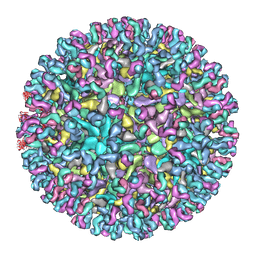 | | Cryo-EM structure of Chikungunya virus in complex with Fab fragments of mAb CHK-124 | | Descriptor: | Capsid protein, E1 glycoprotein, E2 glycoprotein, ... | | Authors: | Zhou, Q.F, Fox, J.M, Earnest, J.T, Ng, T.S, Kim, A.S, Fibriansah, G, Kostyuchenko, V.A, Shu, B, Diamond, M.S, Lok, S.M. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structural basis of Chikungunya virus inhibition by monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4V7G
 
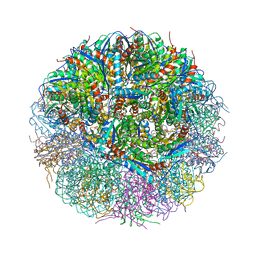 | | Crystal Structure of Lumazine Synthase from Bacillus Anthracis | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Morgunova, E, Illarionov, B, Saller, S, Popov, A, Sambaiah, T, Bacher, A, Cushman, M, Fischer, M, Ladenstein, R. | | Deposit date: | 2009-09-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural study and thermodynamic characterization of inhibitor binding to lumazine synthase from Bacillus anthracis.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
9FWX
 
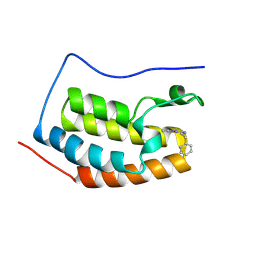 | | Crystal structure of BRD4 BD1 with MEN1404BS. | | Descriptor: | 3-methyl-~{N}-(2-phenylethyl)-[1,2,4]triazolo[4,3-b]pyridazin-6-amine, Bromodomain-containing protein 4 | | Authors: | Bader, G, Kessler, D. | | Deposit date: | 2024-07-01 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Probing Protein-Ligand Methyl-pi Interaction Geometries through Chemical Shift Measurements of Selectively Labeled Methyl Groups.
J.Med.Chem., 2024
|
|
7C5E
 
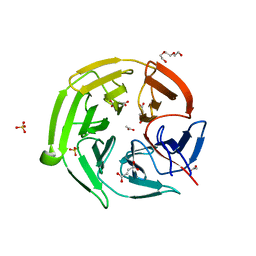 | | Crystal structure of Keap1 in complex with fumarate (FUM) | | Descriptor: | ACETATE ION, FUMARIC ACID, Kelch-like ECH-associated protein 1, ... | | Authors: | Padmanabhan, B, Unni, S, Deshmukh, P. | | Deposit date: | 2020-05-19 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the multiple binding modes of Dimethyl Fumarate (DMF) and its analogs to the Kelch domain of Keap1.
Febs J., 288, 2021
|
|
7C60
 
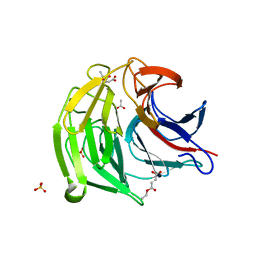 | | Crystal structure of Keap1 in complex with monoethyl fumarate (MEF) | | Descriptor: | (~{Z})-4-ethoxy-4-oxidanylidene-but-2-enoic acid, ACETATE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Padmanabhan, B, Unni, S, Deshmukh, P, Krishnappa, G. | | Deposit date: | 2020-05-21 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the multiple binding modes of Dimethyl Fumarate (DMF) and its analogs to the Kelch domain of Keap1.
Febs J., 288, 2021
|
|
6YEF
 
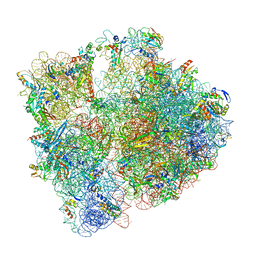 | | 70S initiation complex with assigned rRNA modifications from Staphylococcus aureus | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S11, ... | | Authors: | Fatkhullin, B, Golubev, A, Khusainov, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2020-03-24 | | Release date: | 2021-01-27 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the ribosome functional complex of the human pathogen Staphylococcus aureus at 3.2 angstrom resolution.
Febs Lett., 594, 2020
|
|
6YST
 
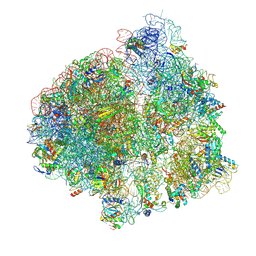 | | Structure of the P+9 ArfB-ribosome complex with P/E hybrid tRNA in the post-hydrolysis state | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
6YSS
 
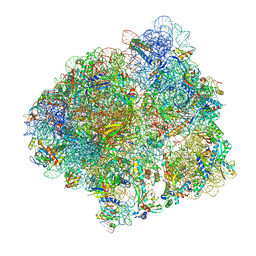 | | Structure of the P+9 ArfB-ribosome complex in the post-hydrolysis state | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
7O9K
 
 | | Human mitochondrial ribosome large subunit assembly intermediate with MTERF4-NSUN4, MRM2, MTG1, the MALSU module, GTPBP5 and mtEF-Tu | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Valentin Gese, G, Hallberg, B.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for late maturation steps of the human mitoribosomal large subunit.
Nat Commun, 12, 2021
|
|
7O9M
 
 | | Human mitochondrial ribosome large subunit assembly intermediate with MTERF4-NSUN4, MRM2, MTG1 and the MALSU module | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Valentin Gese, G, Hallberg, B.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for late maturation steps of the human mitoribosomal large subunit.
Nat Commun, 12, 2021
|
|
6YSU
 
 | | Structure of the P+0 ArfB-ribosome complex in the post-hydrolysis state | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
8EJI
 
 | | Lassa virus glycoprotein complex (Josiah) bound to 19.7E Fab | | Descriptor: | 19.7E Fab heavy chain, 19.7E Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
