7N26
 
 | | NMR structure of EpI-[Y(SO3)15Y]-NH2 | | Descriptor: | Alpha-conotoxin EpI | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N0T
 
 | | NMR structure of EpI[Y(SO)315Y]-OH | | Descriptor: | Alpha-conotoxin EpI | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-26 | | Release date: | 2021-11-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
3T4M
 
 | | Ac-AChBP ligand binding domain mutated to human alpha-7 nAChR (intermediate) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nemecz, A, Taylor, P.W. | | Deposit date: | 2011-07-26 | | Release date: | 2011-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Creating an alpha-7 nicotinic acetylcholine recognition domain from the
acetylcholine binding protein: crystallographic and ligand selectivity analyses
J.Biol.Chem., 286, 2011
|
|
3SH1
 
 | | Ac-AChBP ligand binding domain mutated to human alpha-7 nAChR | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nemecz, A, Taylor, P.W. | | Deposit date: | 2011-06-15 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Creating an alpha-7 nicotinic acetylcholine recognition domain from the
acetylcholine binding protein: crystallographic and ligand selectivity analyses
J.Biol.Chem., 286, 2011
|
|
3SIO
 
 | | Ac-AChBP ligand binding domain (not including beta 9-10 linker) mutated to human alpha-7 nAChR | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nemecz, A, Taylor, P.W. | | Deposit date: | 2011-06-19 | | Release date: | 2011-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Creating an alpha-7 nicotinic acetylcholine recognition domain from the
acetylcholine binding protein: crystallographic and ligand selectivity analyses
J.Biol.Chem., 286, 2011
|
|
8J97
 
 | | Structure of Muscarinic receptor (M2R) in complex with beta-arrestin1 (Local refine, cross-linked) | | Descriptor: | Beta-arrestin-1, Fab30 Heavy Chain, Fab30 Light Chain, ... | | Authors: | Maharana, J, Sano, F.K, Shihoya, W, Banerjee, R, Nureki, O, Shukla, A.K. | | Deposit date: | 2023-05-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
8JAF
 
 | | Structure of Muscarinic receptor (M2R) in complex with beta-arrestin1 (Local Refine, non-cross linked) | | Descriptor: | Beta-arrestin-1, Fab30 heavy chain, Fab30 light chain, ... | | Authors: | Maharana, J, Sano, F.K, Shihoya, W, Banerjee, R, Nureki, O, Shukla, A.K. | | Deposit date: | 2023-05-05 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
5FJV
 
 | | Crystal structure of the extracellular domain of alpha2 nicotinic acetylcholine receptor in pentameric assembly | | Descriptor: | EPIBATIDINE, NEURONAL ACETYLCHOLINE RECEPTOR SUBUNIT ALPHA-2 | | Authors: | Giastas, P, Kouvatsos, N, Chroni-Tzartou, D, Tzartos, S.J. | | Deposit date: | 2015-10-13 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of a Human Neuronal Nachr Extracellular Domain in Pentameric Assembly: Ligand-Bound Alpah2 Homopentamer.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
9BAF
 
 | | Solution NMR structure of conofurin-Delta | | Descriptor: | Alpha-conotoxin LvIA | | Authors: | Harvey, P.J, Craik, D.J, Hone, A.J, McIntosh, J.M. | | Deposit date: | 2024-04-04 | | Release date: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Design, Synthesis, and Structure-Activity Relationships of Novel Peptide Derivatives of the Severe Acute Respiratory Syndrome-Coronavirus-2 Spike-Protein that Potently Inhibit Nicotinic Acetylcholine Receptors.
J.Med.Chem., 67, 2024
|
|
3LII
 
 | | Recombinant human acetylcholinesterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, SULFATE ION | | Authors: | Dvir, H, Rosenberry, T, Harel, M, Silman, I, Sussman, J. | | Deposit date: | 2010-01-25 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Acetylcholinesterase: From 3D structure to function.
Chem.Biol.Interact, 187, 2010
|
|
1K64
 
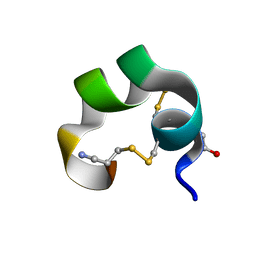 | | NMR Structue of alpha-conotoxin EI | | Descriptor: | alpha-conotoxin EI | | Authors: | Park, K.H, Suk, J.E, Jacobsen, R, Gray, W.R, McIntosh, J.M, Han, K.H. | | Deposit date: | 2001-10-15 | | Release date: | 2003-09-09 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of alpha-conotoxin EI, a neuromuscular toxin specific for the alpha 1/delta subunit interface of torpedo nicotinic acetylcholine receptor
J.BIOL.CHEM., 276, 2001
|
|
3WTM
 
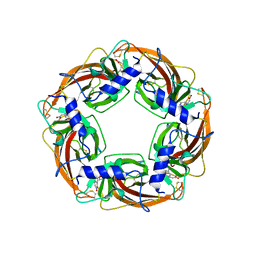 | | Crystal Structure of Lymnaea stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Nitromethylene Analogue of Imidacloprid | | Descriptor: | 2-chloro-5-{[(2E)-2-(nitromethylidene)imidazolidin-1-yl]methyl}pyridine, Acetylcholine-binding protein | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
3WTO
 
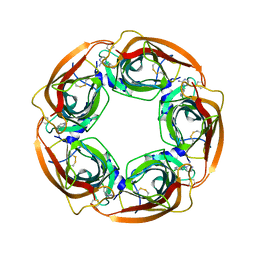 | | Crystal Structure of Lymnaea stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Desnitro-imidacloprid | | Descriptor: | (2Z)-1-[(6-chloropyridin-3-yl)methyl]imidazolidin-2-imine, Acetylcholine-binding protein | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
3WTI
 
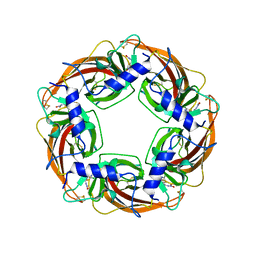 | | Crystal Structure of Lymnaea stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Clothianidin | | Descriptor: | 1-[(2-chloro-1,3-thiazol-5-yl)methyl]-3-methyl-2-nitroguanidine, Acetylcholine-binding protein | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
3WTH
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Imidacloprid | | Descriptor: | (2E)-1-[(6-chloropyridin-3-yl)methyl]-N-nitroimidazolidin-2-imine, Acetylcholine-binding protein | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
3WTL
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine Binding Protein Complexed with Nitromethylene Analogue of Imidacloprid | | Descriptor: | 2-chloro-5-{[(2E)-2-(nitromethylidene)imidazolidin-1-yl]methyl}pyridine, Acetylcholine-binding protein | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
3WTN
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine Binding Protein Complexed with Desnitro-imidacloprid | | Descriptor: | (2Z)-1-[(6-chloropyridin-3-yl)methyl]imidazolidin-2-imine, Acetylcholine-binding protein, CADMIUM ION, ... | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
8FX5
 
 | | Human M4 muscarinic acetylcholine receptor complex with Gi1 and xanomeline | | Descriptor: | Antibody fragment scFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Vuckovic, Z, Mobbs, J.I, Glukhova, A, Sexton, P.M, Danev, R, Thal, D.M. | | Deposit date: | 2023-01-23 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Xanomeline displays concomitant orthosteric and allosteric binding modes at the M 4 mAChR.
Nat Commun, 14, 2023
|
|
2GCZ
 
 | | Solution Structure of alpha-Conotoxin OmIA | | Descriptor: | Alpha-conotoxin OmIA | | Authors: | Chi, S.-W, Kim, D.-H, Olivera, B.M, McIntosh, J.M, Han, K.-H. | | Deposit date: | 2006-03-15 | | Release date: | 2006-07-25 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of a neuronal nicotinic acetylcholine receptor antagonist alpha-conotoxin OmIA that discriminates alpha3 vs. alpha6 nAChR subtypes
Biochem.Biophys.Res.Commun., 345, 2006
|
|
8FLP
 
 | | NMR Solution Structure of LvIC analogue | | Descriptor: | Alpha-conotoxin LvIC analogue | | Authors: | Harvey, P.J, Craik, D.J. | | Deposit date: | 2022-12-22 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-22 | | Method: | SOLUTION NMR | | Cite: | Discovery, Characterization, and Engineering of LvIC, an alpha 4/4-Conotoxin That Selectively Blocks Rat alpha 6/ alpha 3 beta 4 Nicotinic Acetylcholine Receptors.
J.Med.Chem., 66, 2023
|
|
1UL2
 
 | | Solution Conformation of alpha-Conotoxin GIC | | Descriptor: | alpha-conotoxin GIC | | Authors: | Chi, S.-W, Kim, D.-H, Olivera, B.M, McIntosh, J.M, Han, K.-H. | | Deposit date: | 2003-09-06 | | Release date: | 2004-07-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of alpha-conotoxin GIC, a novel potent antagonist of alpha3beta2 nicotinic acetylcholine receptors
Biochem.J., 380, 2004
|
|
1ZLC
 
 | | Solution Conformation of alpha-conotoxin PIA | | Descriptor: | Alpha-conotoxin PIA | | Authors: | Chi, S.-W, Lee, S.-H, Kim, D.-H, Kim, J.-S, Olivera, B.M, McIntosh, J.M, Han, K.-H. | | Deposit date: | 2005-05-05 | | Release date: | 2006-05-02 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of alpha-conotoxin PIA, a novel antagonist of alpha6 subunit containing nicotinic acetylcholine receptors
Biochem.Biophys.Res.Commun., 338, 2005
|
|
2HA4
 
 | | Crystal structure of mutant S203A of mouse acetylcholinesterase complexed with acetylcholine | | Descriptor: | ACETATE ION, ACETYLCHOLINE, Acetylcholinesterase, ... | | Authors: | Bourne, Y, Radic, Z, Sulzenbacher, G, Kim, E, Taylor, P, Marchot, P. | | Deposit date: | 2006-06-12 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Substrate and product trafficking through the active center gorge of acetylcholinesterase analyzed by crystallography and equilibrium binding
J.Biol.Chem., 281, 2006
|
|
2L03
 
 | |
1RGJ
 
 | | NMR STRUCTURE OF THE COMPLEX BETWEEN ALPHA-BUNGAROTOXIN AND MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR WITH ENHANCED ACTIVITY | | Descriptor: | MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR, long neurotoxin 1 | | Authors: | Bernini, A, Spiga, O, Ciutti, A, Scarselli, M, Bracci, L, Lozzi, L, Lelli, B, Neri, P, Niccolai, N. | | Deposit date: | 2003-11-12 | | Release date: | 2003-11-25 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR and MD studies on the interaction between ligand peptides and alpha-bungarotoxin.
J.Mol.Biol., 339, 2004
|
|
