2R32
 
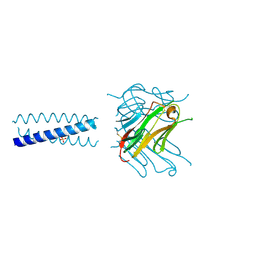 | | Crystal Structure of human GITRL variant | | Descriptor: | GCN4-pII/Tumor necrosis factor ligand superfamily member 18 fusion protein, SULFATE ION | | Authors: | Chattopadhyay, K, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-08-28 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Assembly and structural properties of glucocorticoid-induced TNF receptor ligand: Implications for function.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2G7G
 
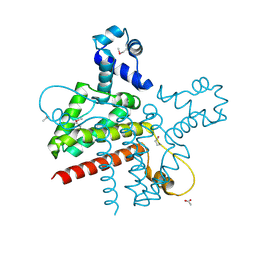 | | The Crystal Structure of the Putative Transcriptional Regulator Rha04620 from Rhodococcus sp. RHA1 | | Descriptor: | ACETIC ACID, Rha04620, Putative Transcriptional Regulator | | Authors: | Kim, Y, Joachimiak, A, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A.M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-28 | | Release date: | 2006-03-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Crystal Structure of the Putative Transcriptional Regulator Rha04620 from Rhodococcus sp. RHA1
To be Published
|
|
2QLZ
 
 | |
4ALV
 
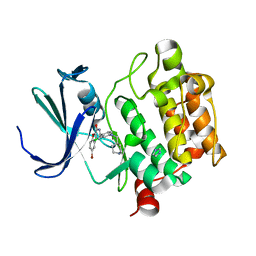 | | Benzofuropyrimidinone Inhibitors of Pim-1 | | Descriptor: | 8-bromo-2-{2-chloro-4-[(piperidin-4-ylmethyl)amino]phenyl}[1]benzofuro[3,2-d]pyrimidin-4(3H)-one, IMIDAZOLE, SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Stout, T.J, Adams, L. | | Deposit date: | 2012-03-05 | | Release date: | 2013-01-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | The Design, Synthesis, and Biological Evaluation of Pim Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3U2R
 
 | |
2QUF
 
 | | Crystal Structure of Transcription Factor AXXA-PF0095 from Pyrococcus furiosus | | Descriptor: | GLYCEROL, Transcription Factor PF0095 | | Authors: | Yang, H, Lipscomb, G.L, Scott, R.A, Rose, J.P, Wang, B.C. | | Deposit date: | 2007-08-05 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Transcription Factor AXXA-PF0095 from Pyrococcus furiosus
To be Published
|
|
2QNP
 
 | | HIV-1 Protease in complex with a iodo decorated pyrrolidine-based inhibitor | | Descriptor: | CHLORIDE ION, Gag-Pol polyprotein (Pr160Gag-Pol), N,N'-(3S,4S)-pyrrolidine-3,4-diylbis[N-(4-iodobenzyl)benzenesulfonamide] | | Authors: | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2007-07-19 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structure-Guided Design of C2-Symmetric HIV-1 Protease Inhibitors Based on a Pyrrolidine Scaffold.
J.Med.Chem., 51, 2008
|
|
4AVB
 
 | | Crystal structure of protein lysine acetyltransferase Rv0998 in complex with acetyl CoA and cAMP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ACETYL COENZYME *A, ... | | Authors: | Lee, H.J, Lang, P.T, Fortune, S.M, Sassetti, C.M, Alber, T. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic AMP Regulation of Protein Lysine Acetylation in Mycobacterium Tuberculosis.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2R2V
 
 | |
2QNN
 
 | | HIV-1 protease in complex with a multiple decorated pyrrolidine-based inhibitor | | Descriptor: | 4,4'-[(3S,4S)-pyrrolidine-3,4-diylbis({[4-(trifluoromethyl)benzyl]imino}sulfonyl)]dibenzamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2007-07-19 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure-Guided Design of C2-Symmetric HIV-1 Protease Inhibitors Based on a Pyrrolidine Scaffold.
J.Med.Chem., 51, 2008
|
|
4AVC
 
 | | Crystal structure of protein lysine acetyltransferase Rv0998 in complex with acetyl CoA and cAMP | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Lee, H.J, Lang, P.T, Fortune, S.M, Sassetti, C.M, Alber, T. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-11 | | Last modified: | 2012-08-22 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Cyclic AMP Regulation of Protein Lysine Acetylation in Mycobacterium Tuberculosis.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VC4
 
 | |
3VBX
 
 | |
2H68
 
 | | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex | | Descriptor: | WD-repeat protein 5 | | Authors: | Ruthenburg, A.J, Wang, W.-K, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-30 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2H6K
 
 | | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex | | Descriptor: | Histone H3 K4-Me 9-residue peptide, WD-repeat protein 5 | | Authors: | Ruthenburg, A.J, Wang, W.-K, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-31 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.
Nat.Struct.Mol.Biol., 13, 2006
|
|
4IA9
 
 | | Crystal structure of human WD REPEAT DOMAIN 5 in complex with 2-chloro-4-fluoro-3-methyl-N-[2-(4-methylpiperazin-1-yl)-5-nitrophenyl]benzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-4-fluoro-3-methyl-N-[2-(4-methylpiperazin-1-yl)-5-nitrophenyl]benzamide, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Dombrovski, L, Bolshan, Y, Getlik, M, Tempel, W, Kuznetsova, E, Wasney, G.A, Hajian, T, Poda, G, Nguyen, K.T, Schapira, M, Brown, P.J, Al-awar, R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Smil, D, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-06 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Synthesis, Optimization, and Evaluation of Novel Small Molecules as Antagonists of WDR5-MLL Interaction.
ACS Med Chem Lett, 4, 2013
|
|
2H9L
 
 | | WDR5delta23 | | Descriptor: | SULFATE ION, WD-repeat protein 5 | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Martin, F, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-10 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for molecular recognition and presentation of histone H3 By WDR5.
Embo J., 25, 2006
|
|
2H6Q
 
 | | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex | | Descriptor: | Histone H3 K4-Me3 9-residue peptide, WD-repeat protein 5 | | Authors: | Ruthenburg, A.J, Wang, W.-K, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-06-01 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.
Nat.Struct.Mol.Biol., 13, 2006
|
|
4HYL
 
 | | The crystal structure of an anti-sigma-factor antagonist from Haliangium ochraceum DSM 14365 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Stage II sporulation protein | | Authors: | Tan, K, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-13 | | Release date: | 2012-11-28 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | The crystal structure of an anti-sigma-factor antagonist from Haliangium ochraceum DSM 14365
To be Published
|
|
3R04
 
 | | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors | | Descriptor: | 5-{6-[(trans-4-aminocyclohexyl)amino]pyrazin-2-yl}-1-benzofuran-2-carboxylic acid, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Xiang, Y, Hirth, B, Asmussen, G, Biemann, H.-P, Good, A, Fitzgerald, M, Gladysheva, T, Jancsics, K, Liu, J, Metz, M, Papoulis, A, Skerlj, R, Stepp, D.J, Wei, R.R. | | Deposit date: | 2011-03-07 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2H14
 
 | |
3UIX
 
 | |
3UMW
 
 | | Crystal structure of Pim1 kinase in complex with inhibitor (Z)-2-[(1H-indazol-3-yl)methylene]-6-methoxy-7-(piperazin-1-ylmethyl)benzofuran-3(2H)-one | | Descriptor: | (2Z)-2-(1H-indazol-3-ylmethylidene)-6-methoxy-7-(piperazin-1-ylmethyl)-1-benzofuran-3(2H)-one, GLYCEROL, Proto-oncogene serine/threonine-protein kinase pim-1, ... | | Authors: | Parker, L.J, Handa, N, Yokoyama, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Rational evolution of a novel type of potent and selective proviral integration site in Moloney murine leukemia virus kinase 1 (PIM1) inhibitor from a screening-hit compound.
J.Med.Chem., 55, 2012
|
|
4KMA
 
 | |
2RHU
 
 | |
