1VTT
 
 | | GT Wobble Base-Pairing in Z-DNA at 1.0 Angstrom Atomic Resolution: The Crystal Structure of d(CGCGTG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*G)-3') | | Authors: | Ho, P.S, Frederick, C.A, Quigley, G.J, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | GT Wobble Base-Pairing in Z-DNA at 1.0 Angstrom Atomic Resolution: The Crystal Structure of d(CGCGTG)
Embo J., 4, 1985
|
|
1VTI
 
 | | DNA-DRUG INTERACTIONS: THE CRYSTAL STRUCTURES OF D(TGATCA) COMPLEXED WITH DAUNOMYCIN | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*TP*GP*TP*AP*CP*A)-3') | | Authors: | Nunn, C.M, Van Meervelt, L, Zhang, S, Moore, M.H, Kennard, O. | | Deposit date: | 1992-03-01 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | DNA-Drug Interactions: The Crystal Structures of d(TGTACA) and d(TGATCA) Complexed with Daunomycin
J.Mol.Biol., 222, 1991
|
|
1VTW
 
 | | AT Base Pairs Are Less Stable than GC Base Pairs in Z-DNA: The Crystal Structure of D(M(5)CGTAM(5)CG) | | Descriptor: | DNA (5'-D(*(CH3)CP*GP*TP*AP*(CH3)CP*G)-3') | | Authors: | Wang, A.H.-J, Hakoshima, T, Van Der Marel, G.A, Van Boom, J.H, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | AT Base Pairs Are Less Stable than GC Base Pairs in Z-DNA: The Crystal Structure of d(m(5)CGTAm(5)CG)
Cell(Cambridge,Mass.), 37, 1984
|
|
2RPH
 
 | | RecT-bound ssDNA | | Descriptor: | DNA (5'-D(*DTP*DAP*DCP*DG)-3') | | Authors: | Masuda, T, Ito, Y, Shibata, T, Mikawa, T. | | Deposit date: | 2008-05-15 | | Release date: | 2009-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A non-canonical DNA structure enables homologous recombination in various genetic systems
J.Biol.Chem., 284, 2009
|
|
2RPE
 
 | | hsRad51-bound ssDNA | | Descriptor: | DNA (5'-D(*DTP*DAP*DCP*DG)-3') | | Authors: | Masuda, T, Ito, Y, Shibata, T, Mikawa, T. | | Deposit date: | 2008-05-15 | | Release date: | 2009-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A non-canonical DNA structure enables homologous recombination in various genetic systems
J.Biol.Chem., 284, 2009
|
|
3PTA
 
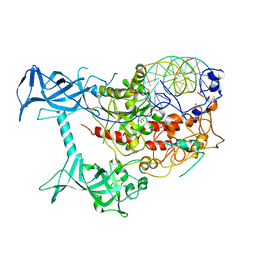 | | Crystal structure of human DNMT1(646-1600) in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*CP*GP*GP*AP*GP*GP*CP*TP*CP*AP*CP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*GP*TP*GP*AP*GP*CP*CP*TP*CP*CP*GP*CP*AP*GP*G)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
1VT9
 
 | |
6AJO
 
 | | Complex form of Uracil DNA glycosylase X and uracil-DNA. | | Descriptor: | DNA (5'-D(P*(ORP)P*TP*T)-3'), IRON/SULFUR CLUSTER, PHOSPHATE ION, ... | | Authors: | Ahn, W.C, Aroli, S, Varshney, U, Woo, E.J. | | Deposit date: | 2018-08-28 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Covalent binding of uracil DNA glycosylase UdgX to abasic DNA upon uracil excision.
Nat.Chem.Biol., 15, 2019
|
|
1VTD
 
 | | UNUSUAL HELICAL PACKING IN CRYSTALS OF DNA BEARING A MUTATION HOT SPOT | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*GP*CP*GP*CP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*GP*CP*GP*CP*CP*GP*GP*T)-3') | | Authors: | Timsit, Y, Westhof, E, Fuchs, R.P.P, Moras, D. | | Deposit date: | 1996-12-12 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unusual helical packing in crystals of DNA bearing a mutation hot spot.
Nature, 341, 1989
|
|
1VAS
 
 | | ATOMIC MODEL OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME COMPLEXED WITH A DNA SUBSTRATE: STRUCTURAL BASIS FOR DAMAGED DNA RECOGNITION | | Descriptor: | DNA (5'-D(*AP*TP*CP*GP*CP*GP*TP*TP*GP*CP*GP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*CP*GP*CP*AP*AP*CP*GP*CP*GP*A)-3'), PROTEIN (T4 ENDONUCLEASE V (E.C.3.1.25.1)) | | Authors: | Vassylyev, D.G, Kashiwagi, T, Mikami, Y, Ariyoshi, M, Iwai, S, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1995-09-08 | | Release date: | 1996-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate: structural basis for damaged DNA recognition.
Cell(Cambridge,Mass.), 83, 1995
|
|
7E15
 
 | |
1VTH
 
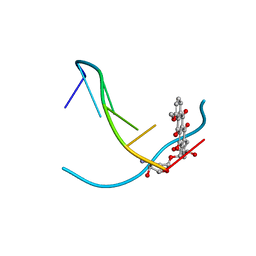 | | DNA-DRUG INTERACTIONS: THE CRYSTAL STRUCTURES OF D(TGTACA) COMPLEXED WITH DAUNOMYCIN | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*TP*GP*TP*AP*CP*A)-3') | | Authors: | Nunn, C.M, Van Meervelt, L, Zhang, S, Moore, M.H, Kennard, O. | | Deposit date: | 1992-03-01 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA-Drug Interactions: The Crystal Structures of d(TGTACA) and d(TGATCA) Complexed with Daunomycin
J.Mol.Biol., 222, 1991
|
|
7Z5A
 
 | |
7Z03
 
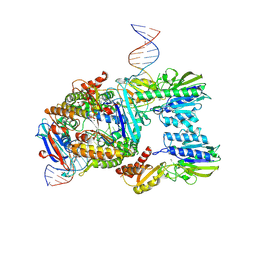 | | Endonuclease state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and extended dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (39-MER), MAGNESIUM ION, ... | | Authors: | Gut, F, Kaeshammer, L, Lammens, K, Bartho, J, van de Logt, E, Kessler, B, Hopfner, K.P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural mechanism of endonucleolytic processing of blocked DNA ends and hairpins by Mre11-Rad50.
Mol.Cell, 82, 2022
|
|
7YZO
 
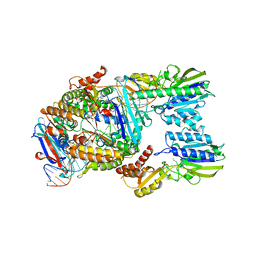 | | Endonuclease state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (31-MER), MAGNESIUM ION, ... | | Authors: | Gut, F, Kaeshammer, L, Lammens, K, Bartho, J, van de Logt, E, Kessler, B, Hopfner, K.P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanism of endonucleolytic processing of blocked DNA ends and hairpins by Mre11-Rad50.
Mol.Cell, 82, 2022
|
|
4ECR
 
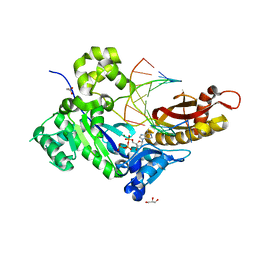 | | Human DNA polymerase eta - DNA ternary complex: Reaction in the AT crystal at pH 7.0 for 40 sec | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Nakamura, T, Zhao, Y, Yang, W. | | Deposit date: | 2012-03-26 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
4ECY
 
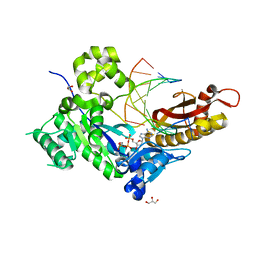 | | Human DNA polymerase eta - DNA ternary complex: AT crystal at pH 6.0 (Na+ MES) with 1 Ca2+ ion | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Nakamura, T, Zhao, Y, Yang, W. | | Deposit date: | 2012-03-26 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
3TQ1
 
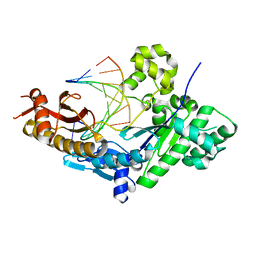 | | Human DNA Polymerase eta in binary complex with DNA | | Descriptor: | DNA (5'-D(*TP*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*TP*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta | | Authors: | Ummat, A, Silverstein, T.D, Jain, R, Buku, A, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2011-09-08 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.556 Å) | | Cite: | Human DNA Polymerase Eta Is Pre-Aligned for dNTP Binding and Catalysis.
J.Mol.Biol., 415, 2012
|
|
4ECV
 
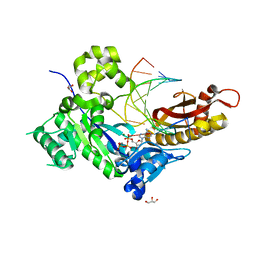 | | Human DNA polymerase eta - DNA ternary complex: Reaction in the AT crystal at pH 7.0 for 230 sec | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Nakamura, T, Zhao, Y, Yang, W. | | Deposit date: | 2012-03-26 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
4ED7
 
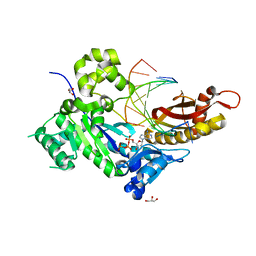 | | Human DNA polymerase eta - DNA ternary complex: TG crystal at pH 7.0 (K+ MES) with 1 Ca2+ ion | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*G)-3'), ... | | Authors: | Nakamura, T, Zhao, Y, Yang, W. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
4ECU
 
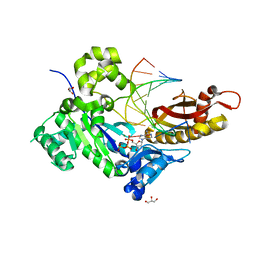 | | Human DNA polymerase eta - DNA ternary complex: Reaction in the AT crystal at pH 7.0 for 200 sec | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Nakamura, T, Zhao, Y, Yang, W. | | Deposit date: | 2012-03-26 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
4ED2
 
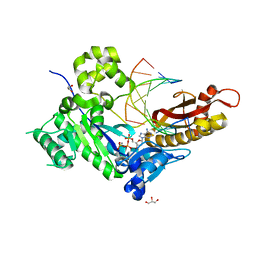 | | Human DNA polymerase eta - DNA ternary complex: AT crystal at pH 7.2 (Na+ HEPES) with 1 Ca2+ ion | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Nakamura, T, Zhao, Y, Yang, W. | | Deposit date: | 2012-03-26 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.711 Å) | | Cite: | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
4ECZ
 
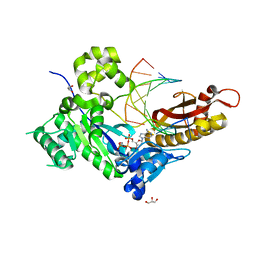 | | Human DNA polymerase eta - DNA ternary complex: AT crystal at pH 6.5 (Na+ MES) with 1 Ca2+ ion | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Nakamura, T, Zhao, Y, Yang, W. | | Deposit date: | 2012-03-26 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.834 Å) | | Cite: | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
4ED8
 
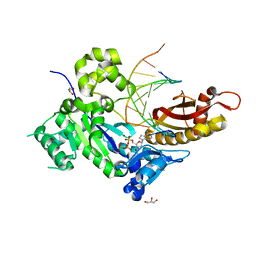 | | Human DNA polymerase eta - DNA ternary complex: Reaction in the TG crystal at pH 7.0, Normal translocation | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*G)-3'), ... | | Authors: | Nakamura, T, Zhao, Y, Yang, W. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
10MH
 
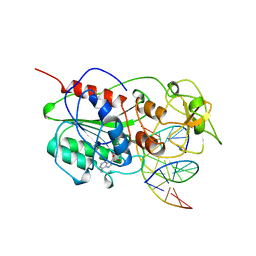 | | TERNARY STRUCTURE OF HHAI METHYLTRANSFERASE WITH ADOHCY AND HEMIMETHYLATED DNA CONTAINING 5,6-DIHYDRO-5-AZACYTOSINE AT THE TARGET | | Descriptor: | DNA (5'-D(P*CP*CP*AP*TP*GP*(5CM)P*GP*CP*TP*GP*AP*C)-3'), DNA (5'-D(P*GP*TP*CP*AP*GP*5NCP*GP*CP*AP*TP*GP*G)-3'), PROTEIN (CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI), ... | | Authors: | Sheikhnejad, G, Brank, A, Christman, J.K, Goddard, A, Alvarez, E, Ford Junior, H, Marquez, V.E, Marasco, C.J, Sufrin, J.R, O'Gara, M, Cheng, X. | | Deposit date: | 1998-08-10 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Mechanism of inhibition of DNA (cytosine C5)-methyltransferases by oligodeoxyribonucleotides containing 5,6-dihydro-5-azacytosine.
J.Mol.Biol., 285, 1999
|
|
