8TYS
 
 | |
8TYQ
 
 | | Structure of the C-terminal half of LRRK2 bound to GZD-824 (G2019S mutant) | | Descriptor: | 4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-3-[(1H-pyrazolo[3,4-b]pyridin-5-yl)ethynyl]benzamide, Designed Ankyrin Repeats Protein E11, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Villagran-Suarez, A, Sanz-Murillo, M, Alegrio-Louro, J, Leschziner, A. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Inhibition of Parkinson's disease-related LRRK2 by type I and type II kinase inhibitors: Activity and structures.
Sci Adv, 9, 2023
|
|
8TYP
 
 | | Complement Protease C1s Inhibited by 6-(4-phenylpiperazin-1-yl)pyridine-3-carboximidamide | | Descriptor: | 6-(4-phenylpiperazin-1-yl)pyridine-3-carboximidamide, Complement C1s subcomponent, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Geisbrecht, B.V. | | Deposit date: | 2023-08-25 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of the C1s Protease and the Classical Complement Pathway by 6-(4-Phenylpiperazin-1-yl)Pyridine-3-Carboximidamide and Chemical Analogs.
J Immunol., 212, 2024
|
|
8TYO
 
 | |
8TYN
 
 | |
8TYM
 
 | |
8TYL
 
 | |
8TYJ
 
 | |
8TYI
 
 | |
8TY7
 
 | |
8TY6
 
 | |
8TY1
 
 | |
8TXZ
 
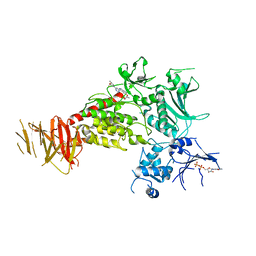 | | Structure of C-terminal LRRK2 bound to MLi-2 | | Descriptor: | (2~{R},6~{S})-2,6-dimethyl-4-[6-[5-(1-methylcyclopropyl)oxy-1~{H}-indazol-3-yl]pyrimidin-4-yl]morpholine, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Sanz-Murillo, M, Villagran-Suarez, A, Alegrio-Louro, J, Leschziner, A. | | Deposit date: | 2023-08-24 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Inhibition of Parkinson's disease-related LRRK2 by type I and type II kinase inhibitors: Activity and structures.
Sci Adv, 9, 2023
|
|
8TXY
 
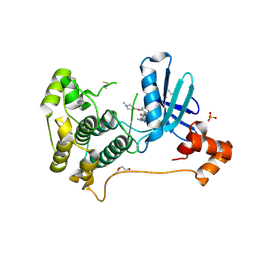 | | X-ray crystal structure of JRD-SIK1/2i-3 bound to a MARK2-SIK2 chimera | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[(5P,8R)-5-(2-cyano-5-{[(3R)-1-methylpyrrolidin-3-yl]methoxy}pyridin-4-yl)pyrazolo[1,5-a]pyridin-2-yl]cyclopropanecarboxamide, SULFATE ION, ... | | Authors: | Raymond, D.D, Lemke, C.T, Shaffer, P.L, Collins, B, Steele, R, Seierstad, M. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of highly selective SIK1/2 inhibitors that modulate innate immune activation and suppress intestinal inflammation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TXX
 
 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A K15 in complex with RNF168 (Class 3) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8TXW
 
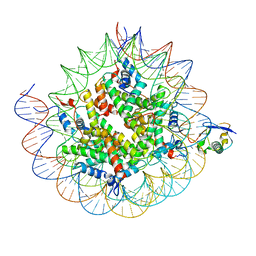 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A K15 in complex with RNF168 (Class 2) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8TXV
 
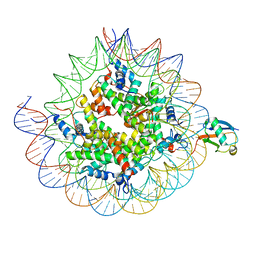 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A K15 in complex with RNF168 (Class 1) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8TXT
 
 | |
8TXS
 
 | |
8TXR
 
 | | E. coli ExoVII(H238A) | | Descriptor: | Exodeoxyribonuclease 7 large subunit, Exodeoxyribonuclease 7 small subunit | | Authors: | Liu, C, Berger, J.M. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of Escherichia coli exonuclease VII.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TXP
 
 | |
8TXO
 
 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound to the unnatural dZ-PTP base pair in the active site | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Aiyer, S, Shan, Z, Oh, J, Wang, D, Tan, Y.Z, Lyumkis, D. | | Deposit date: | 2023-08-23 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Overcoming resolution attenuation during tilted cryo-EM data collection.
Nat Commun, 15, 2024
|
|
8TXN
 
 | |
8TXM
 
 | |
8TXH
 
 | | Crystal structure of KRAS G12D in complex with GDP and compound 14 | | Descriptor: | (4P)-2-amino-4-{4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}-6-(trifluoromethyl)quinazolin-7-yl}-7-fluoro-1-benzothiophene-3-carbonitrile, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P, Irimia, A, Yang, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Based Design and Synthesis of Potent and Selective KRAS G12D Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
