9F1J
 
 | | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 14 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-[2-[2-[2-[2-[2-[2-[2,6-bis(oxidanylidene)piperidin-3-yl]-1,3-bis(oxidanylidene)isoindol-4-yl]oxyethanoylamino]ethoxy]ethoxy]ethoxy]ethyl]-4-[4-[2-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(2-morpholin-4-ylethyl)benzimidazol-2-yl]ethyl]phenoxy]butanamide | | Authors: | Balourdas, D.I, Edmonds, A.K, Marsh, G.P, Maple, H.J, Spencer, J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-11 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (1.13003039 Å) | | Cite: | Structure-Guided Design of ISOX-DUAL-Based Degraders Targeting BRD4 and CBP/EP300: A Case of Degrader Collapse.
J.Med.Chem., 68, 2025
|
|
5DQ1
 
 | | Endothiapepsin in complex with fragment 34 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(cyclopropylamino)methyl]-8-ethylquinolin-2(1H)-one, Endothiapepsin, ... | | Authors: | Radeva, N, Uehlein, M, Weiss, M, Heine, A, Klebe, G. | | Deposit date: | 2015-09-14 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
3MHJ
 
 | | Human tankyrase 2 - catalytic PARP domain in complex with 1-methyl-3-(trifluoromethyl)-5h-benzo[c][1,8]naphtyridine-6-one | | Descriptor: | 1-methyl-3-(trifluoromethyl)benzo[c][1,8]naphthyridin-6(5H)-one, SULFATE ION, Tankyrase-2, ... | | Authors: | Karlberg, T, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kotenyova, T, Markova, N, Moche, M, Nordlund, P, Nyman, T, Persson, C, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-08 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors
Nat.Biotechnol., 30, 2012
|
|
6ZMO
 
 | | SARS-CoV-2 Nsp1 bound to the human LYAR-80S-eEF1a ribosome complex | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6JTU
 
 | | Crystal structure of MHETase from Ideonella sakaiensis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Sagong, H.-Y, Seo, H, Kim, K.-J. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Decomposition of PET film by MHETase using Exo-PETase function
Acs Catalysis, 10, 2020
|
|
8ZWY
 
 | | Structure-Based Mechanism and Specificity of Human Galactosyltransferase B3GalT5 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-methyl beta-D-galactopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lo, J.M, Ma, C. | | Deposit date: | 2024-06-13 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Mechanism and Specificity of Human Galactosyltransferase beta 3GalT5.
J.Am.Chem.Soc., 147, 2025
|
|
7K6M
 
 | | Crystal structure of PI3Kalpha selective Inhibitor PF-06843195 | | Descriptor: | 2,2-difluoroethyl (3S)-3-{[2'-amino-5-fluoro-2-(morpholin-4-yl)[4,5'-bipyrimidin]-6-yl]amino}-3-(hydroxymethyl)pyrrolidine-1-carboxylate, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Chen, P, Brooun, A, Deng, Y.L, Grodsky, N, Kaiser, S.E. | | Deposit date: | 2020-09-21 | | Release date: | 2021-01-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structure-Based Drug Design and Synthesis of PI3K alpha-Selective Inhibitor (PF-06843195).
J.Med.Chem., 64, 2021
|
|
3MHI
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-{[(5-nitro-6-oxo-1,6-dihydro-4-pyrimidinyl)amino]methyl}benzenesulfonamide | | Descriptor: | 4-{[(5-nitro-6-oxo-1,6-dihydropyrimidin-4-yl)amino]methyl}benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Grazulis, S, Manakova, E, Golovenko, D. | | Deposit date: | 2010-04-08 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 4-[N-(Substituted 4-pyrimidinyl)amino]benzenesulfonamides as inhibitors of carbonic anhydrase isozymes I, II, VII, and XIII
Bioorg.Med.Chem., 18, 2010
|
|
4O19
 
 | | The crystal structure of a mutant NAMPT (G217V) | | Descriptor: | 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
8ZX2
 
 | | Structure-Based Mechanism and Specificity of Human Galactosyltransferase B3GalT5 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lo, J.M, Ma, C. | | Deposit date: | 2024-06-13 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Mechanism and Specificity of Human Galactosyltransferase beta 3GalT5.
J.Am.Chem.Soc., 147, 2025
|
|
6JVP
 
 | | Crystal structure of human MTH1 in complex with compound MI1024 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-cyclopropyl-6-piperidin-1-yl-pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
2JDO
 
 | | STRUCTURE OF PKB-BETA (AKT2) COMPLEXED WITH ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY) ETHYLAMINO)ETHYL)AMIDE | | Descriptor: | 1,2-ETHANEDIOL, GLYCOGEN SYNTHASE KINASE-3 BETA, ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY)ETHYLAMINO)ETHYL)AMIDE, ... | | Authors: | Davies, T.G, Verdonk, M.L, Graham, B, Saalau-Bethell, S, Hamlett, C.C.F, Mchardy, T, Collins, I, Garrett, M.D, Workman, P, Woodhead, S.J, Jhoti, H, Barford, D. | | Deposit date: | 2007-01-11 | | Release date: | 2007-02-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Structural Comparison of Inhibitor Binding to Pkb, Pka and Pka-Pkb Chimera
J.Mol.Biol., 367, 2007
|
|
5JDV
 
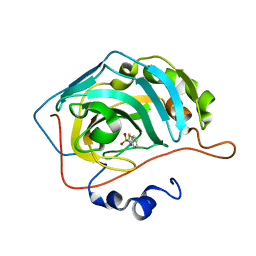 | | Human carbonic anhydrase II (F131W) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-17 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
4O2D
 
 | |
9J0N
 
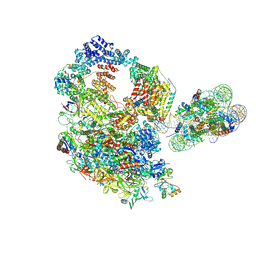 | | Paused elongation complex of mammalian RNA polymerase II with nucleosome (PEC2-nuc) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Naganuma, M, Kujirai, T, Ehara, H, Uejima, T, Ito, T, Goto, M, Aoki, M, Henmi, M, Miyamoto-Kohno, S, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2024-08-02 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into promoter-proximal pausing of RNA polymerase II at +1 nucleosome.
Sci Adv, 11, 2025
|
|
3MNZ
 
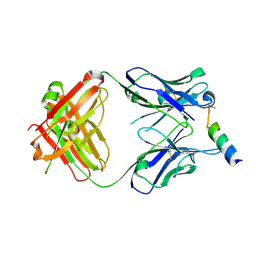 | | Crystal structure of the non-neutralizing HIV antibody 13H11 Fab fragment with a gp41 MPER-derived peptide bearing Ala substitutions in a helical conformation | | Descriptor: | ANTI-HIV-1 ANTIBODY 13H11 HEAVY CHAIN, ANTI-HIV-1 ANTIBODY 13H11 LIGHT CHAIN, SODIUM ION, ... | | Authors: | Nicely, N.I, Dennison, S.M, Kelsoe, G, Liao, H.-X, Alam, S.M, Haynes, B.F. | | Deposit date: | 2010-04-22 | | Release date: | 2010-11-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Non-Neutralizing HIV-1 gp41 Envelope Antibody Demonstrates Neutralization Mechanism of gp41 Antibodies
To be Published
|
|
8A2D
 
 | | EGFR kinase domain (L858R/V948R) in complex with 2-[4-(difluoromethyl)-6-[2-[4-[[4-(hydroxymethyl)-1-piperidyl]methyl]phenyl]ethynyl]-7-methyl-indazol-2-yl]-2-spiro[6,7-dihydropyrrolo[1,2-c]imidazole-5,1'-cyclopropane]-1-yl-N-thiazol-2-yl-acetamide | | Descriptor: | (2R)-2-[4-[bis(fluoranyl)methyl]-6-[2-[4-[[4-(hydroxymethyl)piperidin-1-yl]methyl]phenyl]ethynyl]-7-methyl-indazol-2-yl]-2-spiro[6,7-dihydropyrrolo[1,2-c]imidazole-5,1'-cyclopropane]-1-yl-N-(1,3-thiazol-2-yl)ethanamide, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kuglstatter, A, Ehler, A. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Discovery of Novel Allosteric EGFR L858R Inhibitors for the Treatment of Non-Small-Cell Lung Cancer as a Single Agent or in Combination with Osimertinib.
J.Med.Chem., 65, 2022
|
|
8ZWP
 
 | | Structure-Based Mechanism and Specificity of Human Galactosyltransferase B3GalT5 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lo, J.M, Ma, C. | | Deposit date: | 2024-06-13 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-Based Mechanism and Specificity of Human Galactosyltransferase beta 3GalT5.
J.Am.Chem.Soc., 147, 2025
|
|
3MQP
 
 | | Crystal Structure of human BFL-1 in complex with NOXA BH3 peptide, Northeast Structural Genomics Consortium Target HR2930 | | Descriptor: | Bcl-2-related protein A1, Phorbol-12-myristate-13-acetate-induced protein 1 | | Authors: | Guan, R, Xiao, R, Zhao, L, Acton, T.B, Gelinas, C, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of human BFL-1 in complex with NOXA BH3 peptide
To be Published
|
|
7TDU
 
 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-1 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxo(1-~2~H)pyrrolidin-3-yl]propan-2-yl}-3-{N-[tert-butyl(~2~H)carbamoyl]-3-methyl-L-(N-~2~H)valyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-(~2~H)carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
8ZWW
 
 | | Structure-Based Mechanism and Specificity of Human Galactosyltransferase B3GalT5 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lo, J.M, Ma, C. | | Deposit date: | 2024-06-13 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-Based Mechanism and Specificity of Human Galactosyltransferase beta 3GalT5.
J.Am.Chem.Soc., 147, 2025
|
|
5JGA
 
 | | Crystal structure of human TAK1/TAB1 fusion protein in complex with ligand 11c | | Descriptor: | N-[5-(4-methylpiperazine-1-carbonyl)[1,1'-biphenyl]-2-yl]-4-oxo-3,4-dihydrothieno[3,2-d]pyrimidine-7-carboxamide, TAK1 kinase - TAB1 chimera fusion protein | | Authors: | Irie, M, Nakamura, M, Fukami, T.A, Matsuura, T, Morishima, K. | | Deposit date: | 2016-04-19 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a potent and highly selective transforming growth factor beta receptor-associated kinase 1 (TAK1) inhibitor by structure based drug design (SBDD)
Bioorg.Med.Chem., 24, 2016
|
|
1M7Q
 
 | | Crystal structure of p38 MAP kinase in complex with a dihydroquinazolinone inhibitor | | Descriptor: | 1-(2,6-DICHLOROPHENYL)-5-(2,4-DIFLUOROPHENYL)-7-PIPERAZIN-1-YL-3,4-DIHYDROQUINAZOLIN-2(1H)-ONE, Mitogen-activated protein kinase 14, SULFATE ION | | Authors: | Stelmach, J.E, Liu, L, Patel, S.B, Pivnichny, J.V, Scapin, G, Singh, S, Hop, C.E.C.A, Wang, Z, Cameron, P.M, Nichols, E.A, O'Keefe, S.J, O'Neill, E.A, Schmatz, D.M, Schwartz, C.D, Thompson, C.M, Zaller, D.M, Doherty, J.B. | | Deposit date: | 2002-07-22 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of potent, orally bioavailable dihydroquinazolinone inhibitors of p38 MAP kinase.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
2W1S
 
 | |
6UO5
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 1 (CD1) Y363F mutant complexed with AR-42 | | Descriptor: | 1,2-ETHANEDIOL, AR-42, Histone deacetylase 6, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2019-10-14 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.43934226 Å) | | Cite: | Structural Basis of Catalysis and Inhibition of HDAC6 CD1, the Enigmatic Catalytic Domain of Histone Deacetylase 6.
Biochemistry, 58, 2019
|
|
