3U8M
 
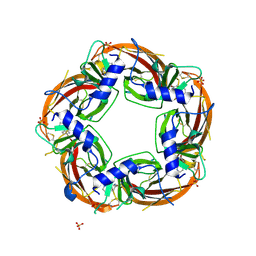 | | Crystal structure of the acetylcholine binding protein (AChBP) from Lymnaea stagnalis in complex with NS3920 (1-(6-bromopyridin-3-yl)-1,4-diazepane) | | Descriptor: | 1-(6-bromopyridin-3-yl)-1,4-diazepane, Acetylcholine-binding protein, SULFATE ION | | Authors: | Rohde, L.A.H, Ahring, P.K, Jensen, M.L, Nielsen, E.O, Peters, D, Helgstrand, C, Krintel, C, Harpsoe, K, Gajhede, M, Kastrup, J.S, Balle, T. | | Deposit date: | 2011-10-17 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Intersubunit bridge formation governs agonist efficacy at nicotinic acetylcholine alpha 4 beta 2 receptors: unique role of halogen bonding revealed.
J.Biol.Chem., 287, 2012
|
|
2WQ7
 
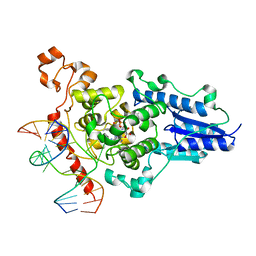 | | Structure of the 6-4 photolyase of D. melanogaster in complex with the non-natural N4-methyl T(6-4)C lesion | | Descriptor: | 5'-D(*AP*CP*AP*GP*CP*GP*GP*TDYP*ZP*GP* CP*AP*AP*GP*T)-3', 5'-D(*TP*AP*CP*CP*TP*GP*CP*GP*AP*CP* CP*GP*CP*TP*G)-3', FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Glas, A.F, Kaya, E, Schneider, S, Maul, M.J, Carell, T. | | Deposit date: | 2009-08-14 | | Release date: | 2010-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA (6-4) Photolyases Reduce Dewar Isomers for Isomerization Into (6-4) Lesions.
J.Am.Chem.Soc., 132, 2010
|
|
1EJC
 
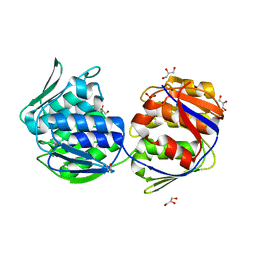 | | Crystal structure of unliganded mura (type2) | | Descriptor: | GLYCEROL, PHOSPHATE ION, UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYLTRANSFERASE | | Authors: | Eschenburg, S, Schonbrunn, E. | | Deposit date: | 2000-03-02 | | Release date: | 2000-10-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparative X-ray analysis of the un-liganded fosfomycin-target murA.
Proteins, 40, 2000
|
|
3GLL
 
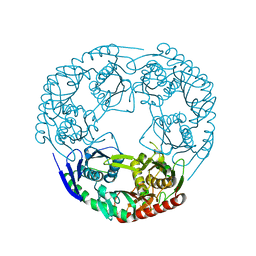 | | Crystal structure of Polynucleotide Phosphorylase (PNPase) core | | Descriptor: | Polyribonucleotide nucleotidyltransferase | | Authors: | Nurmohamed, S, Luisi, B.L. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Escherichia coli polynucleotide phosphorylase core bound to RNase E, RNA and manganese: implications for catalytic mechanism and RNA degradosome assembly.
J.Mol.Biol., 389, 2009
|
|
5A2M
 
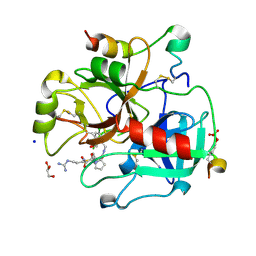 | | Thrombin Inhibitor | | Descriptor: | (2S)-1-[(2R)-5-carbamimidamido-2-[(phenylmethyl)sulfonylamino]pentanoyl]-N-[[5-chloranyl-2-(hydroxymethyl)phenyl]methyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2015-05-20 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Thrombin Inhibition
To be Published
|
|
1AQW
 
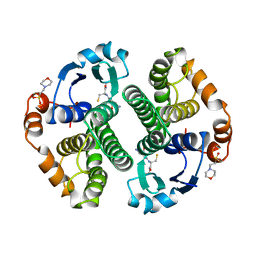 | | GLUTATHIONE S-TRANSFERASE IN COMPLEX WITH GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Prade, L, Huber, R, Manoharan, T.H, Fahl, W.E, Reuter, W. | | Deposit date: | 1997-08-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of class pi glutathione S-transferase from human placenta in complex with substrate, transition-state analogue and inhibitor.
Structure, 5, 1997
|
|
1LJT
 
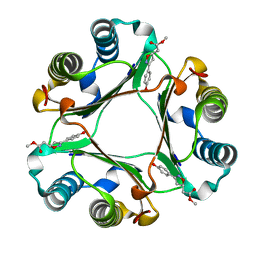 | | Crystal Structure of Macrophage Migration Inhibitory Factor complexed with (S,R)-3-(4-hydroxyphenyl)-4,5-dihydro-5-isoxazole-acetic acid methyl ester (ISO-1) | | Descriptor: | 3-(4-HYDROXYPHENYL)-4,5-DIHYDRO-5-ISOXAZOLE-ACETIC ACID METHYL ESTER, Macrophage migration inhibitory factor | | Authors: | Lubetsky, J.B, Dios, A, Han, J, Aljabari, B, Ruzsicska, B, Mitchell, R, Lolis, E, Al-Abed, Y. | | Deposit date: | 2002-04-22 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Tautomerase Active Site of Macrophage Migration Inhibitory factor is a Potential Target for Discovery of Novel Anti-inflammatory Agents
J.Biol.Chem., 277, 2002
|
|
1LKA
 
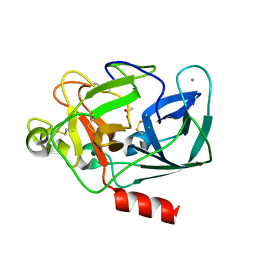 | | Porcine Pancreatic Elastase/Ca-Complex | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Weiss, M.S, Panjikar, S, Nowak, E, Tucker, P.A. | | Deposit date: | 2002-04-24 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal binding to porcine pancreatic elastase: calcium or not calcium.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1O0D
 
 | | Human Thrombin complexed with a d-Phe-Pro-Arg-type Inhibitor and a C-terminal Hirudin derived exo-site inhibitor | | Descriptor: | (2-{2-[(5-CARBAMIMIDOYL-1-METHYL-1H-PYRROL-2-YLMETHYL)-CARBAMOYL]-PYRROL-1-YL}- 1-CYCLOHEXYLMETHYL-2-OXO-ETHYLAMINO)-ACETIC ACID, Decapeptide Hirudin Analogue, Thrombin heavy chain, ... | | Authors: | Lange, U.E, Bauke, D, Hornberger, W, Mack, H, Seitz, W, Hoeffken, H.W. | | Deposit date: | 2003-02-21 | | Release date: | 2003-10-14 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | D-Phe-Pro-Arg type thrombin inhibitors: unexpected selectivity by modification of the P1 moiety
Bioorg.Med.Chem.Lett., 13, 2003
|
|
3NEH
 
 | | Crystal structure of the protein LMO2462 from Listeria monocytogenes complexed with ZN and phosphonate mimic of dipeptide L-Leu-D-Ala | | Descriptor: | (2R)-3-[(R)-[(1R)-1-amino-3-methylbutyl](hydroxy)phosphoryl]-2-methylpropanoic acid, Renal dipeptidase family protein, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-08 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Crystal structure of the protein LMO2462 from Listeria monocytogenes
complexed with ZN and phosphonate mimic of dipeptide L-Leu-D-Ala
To be Published
|
|
1L5T
 
 | | Crystal Structure of a Domain-Opened Mutant (R121D) of the Human Lactoferrin N-lobe Refined From a Merohedrally-Twinned Crystal Form. | | Descriptor: | lactoferrin | | Authors: | Jameson, G.B, Anderson, B.F, Breyer, W.A, Tweedie, J.W, Baker, E.N. | | Deposit date: | 2002-03-07 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a domain-opened mutant (R121D) of the human lactoferrin N-lobe refined from a merohedrally twinned crystal form.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3N7R
 
 | | Crystal structure of the ectodomain complex of the CGRP receptor, a Class-B GPCR, reveals the site of drug antagonism | | Descriptor: | Calcitonin gene-related peptide type 1 receptor, N-[(3R,6S)-6-(2,3-difluorophenyl)-2-oxo-1-(2,2,2-trifluoroethyl)azepan-3-yl]-4-(2-oxo-2,3-dihydro-1H-imidazo[4,5-b]pyridin-1-yl)piperidine-1-carboxamide, N-{(1S)-5-amino-1-[(4-pyridin-4-ylpiperazin-1-yl)carbonyl]pentyl}-3,5-dibromo-Nalpha-{[4-(2-oxo-1,4-dihydroquinazolin-3 (2H)-yl)piperidin-1-yl]carbonyl}-D-tyrosinamide, ... | | Authors: | Ter Haar, E. | | Deposit date: | 2010-05-27 | | Release date: | 2010-09-15 | | Last modified: | 2022-10-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Ectodomain Complex of the CGRP Receptor, a Class-B GPCR, Reveals the Site of Drug Antagonism.
Structure, 18, 2010
|
|
1Y39
 
 | | Co-evolution of protein and RNA structures within a highly conserved ribosomal domain | | Descriptor: | 50S ribosomal protein L11, 58 Nucleotide Ribosomal 23S RNA Domain, COBALT (III) ION, ... | | Authors: | Dunstan, M.S, GuhaThakurta, D, Draper, D.E, Conn, G.L. | | Deposit date: | 2004-11-24 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Coevolution of Protein and RNA Structures within a Highly Conserved Ribosomal Domain
Chem.Biol., 12, 2005
|
|
4ZRT
 
 | | PTP1BC215S bound to Nephrin peptide substrate | | Descriptor: | CHLORIDE ION, GLY-PRO-LEU-PTR-ASP-GLU, GLYCEROL, ... | | Authors: | Selner, N.G, Bell, C.E, Pei, D. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Diverse levels of sequence selectivity and catalytic efficiency of protein-tyrosine phosphatases.
Biochemistry, 53, 2014
|
|
1AZV
 
 | | FAMILIAL ALS MUTANT G37R CUZNSOD (HUMAN) | | Descriptor: | COPPER (II) ION, COPPER/ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Liu, H, Pellegrini, M, Nersissian, A.M, Gralla, E.B, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1997-11-21 | | Release date: | 1998-02-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Subunit asymmetry in the three-dimensional structure of a human CuZnSOD mutant found in familial amyotrophic lateral sclerosis.
Protein Sci., 7, 1998
|
|
3NA8
 
 | | Crystal Structure of a putative dihydrodipicolinate synthetase from Pseudomonas aeruginosa | | Descriptor: | D-MALATE, MAGNESIUM ION, putative dihydrodipicolinate synthetase | | Authors: | Qiu, W, Lam, R, Romanov, V, Jones, K, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2010-06-01 | | Release date: | 2011-06-01 | | Last modified: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of a putative dihydrodipicolinate synthetase from Pseudomonas aeruginosa
To be Published
|
|
6I1R
 
 | | Crystal structure of CMP bound CST in an outward facing conformation | | Descriptor: | CMP-sialic acid transporter 1, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Nji, E, Gulati, A, Qureshi, A.A, Drew, D. | | Deposit date: | 2018-10-30 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the delivery of activated sialic acid into Golgi for sialyation.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6I21
 
 | | Flavin Analogue Sheds Light on Light-Oxygen-Voltage Domain Mechanism | | Descriptor: | 1,2-ETHANEDIOL, Aureochrome1-like protein, CHLORIDE ION, ... | | Authors: | Rizkallah, P.J, Kalvaitis, M.E, Allemann, R.K, Mart, R.J, Johnson, L.A. | | Deposit date: | 2018-10-31 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Noncanonical Chromophore Reveals Structural Rearrangements of the Light-Oxygen-Voltage Domain upon Photoactivation.
Biochemistry, 58, 2019
|
|
2GU2
 
 | | Crystal Structure of an Aspartoacylase from Rattus norvegicus | | Descriptor: | Aspa protein, SULFATE ION, ZINC ION | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-28 | | Release date: | 2006-06-20 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structure of aspartoacylase, the brain enzyme impaired in Canavan disease.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3U8L
 
 | | Crystal structure of the acetylcholine binding protein (AChBP) from Lymnaea stagnalis in complex with NS3570 (1-(5-phenylpyridin-3-yl)-1,4-diazepane) | | Descriptor: | 1-(5-phenylpyridin-3-yl)-1,4-diazepane, Acetylcholine-binding protein, SULFATE ION | | Authors: | Rohde, L.A.H, Ahring, P.K, Jensen, M.L, Nielsen, E.O, Peters, D, Helgstrand, C, Krintel, C, Harpsoe, K, Gajhede, M, Kastrup, J.S, Balle, T. | | Deposit date: | 2011-10-17 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Intersubunit bridge formation governs agonist efficacy at nicotinic acetylcholine alpha 4 beta 2 receptors: unique role of halogen bonding revealed.
J.Biol.Chem., 287, 2012
|
|
6I24
 
 | | Flavin Analogue Sheds Light on Light-Oxygen-Voltage Domain Mechanism | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-5-O-phosphono-D-ribitol, ACETATE ION, ... | | Authors: | Rizkallah, P.J, Kalvaitis, M.E, Allemann, R.K, Mart, R.J, Johnson, L.A. | | Deposit date: | 2018-10-31 | | Release date: | 2019-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A Noncanonical Chromophore Reveals Structural Rearrangements of the Light-Oxygen-Voltage Domain upon Photoactivation.
Biochemistry, 58, 2019
|
|
2X1W
 
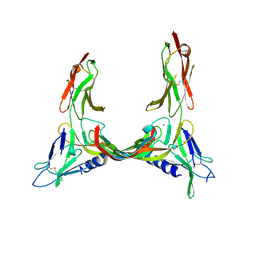 | | Crystal Structure of VEGF-C in Complex with Domains 2 and 3 of VEGFR2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CESIUM ION, ... | | Authors: | Leppanen, V.M, Prota, A.E, Jeltsch, M, Anisimov, A, Kalkkinen, N, Strandin, T, Lankinen, H, Goldman, A, Ballmer-Hofer, K, Alitalo, K. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Determinants of Growth Factor Binding and Specificity by Vegf Receptor 2.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1Y1M
 
 | |
5DCB
 
 | |
1Y2V
 
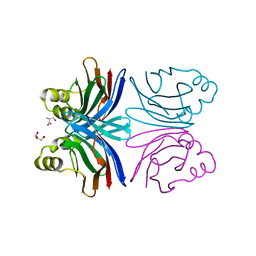 | | Crystal structure of the common edible mushroom (Agaricus bisporus) lectin in complex with T-antigen | | Descriptor: | SERINE, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, lectin | | Authors: | Carrizo, M.E, Capaldi, S, Perduca, M, Irazoqui, F.J, Nores, G.A, Monaco, H.L. | | Deposit date: | 2004-11-23 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Antineoplastic Lectin of the Common Edible Mushroom (Agaricus bisporus) Has Two Binding Sites, Each Specific for a Different Configuration at a Single Epimeric Hydroxyl
J.Biol.Chem., 280, 2005
|
|
