1MCD
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | Immunoglobulin lambda-1 light chain, PEPTIDE N-ACETYL-D-PHE-B-ALA-L-HIS-D-PRO-NH2 | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
1MCJ
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | ACETYL GROUP, AMINO GROUP, D-PHENYLALANINE, ... | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
1MCK
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | IMMUNOGLOBULIN LAMBDA DIMER MCG (LIGHT CHAIN), PEPTIDE N-ACETYL-D-GLU-L-HIS-D-PRO-NH2 | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
1MCF
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | Immunoglobulin lambda-1 light chain, PEPTIDE N-ACETYL-L-GLN-D-PHE-L-HIS-D-PRO-B-ALA-B-ALA-OH | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
1IUH
 
 | | Crystal structure of TT0787 of thermus thermophilus HB8 | | Descriptor: | 2'-5' RNA Ligase | | Authors: | Kato, M, Sakai, H, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-05 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the 2'-5' RNA Ligase from Thermus thermophilus HB8
J.MOL.BIOL., 329, 2003
|
|
1PBQ
 
 | |
3OVE
 
 | |
3S8N
 
 | |
3S8O
 
 | |
3DPL
 
 | |
1MXU
 
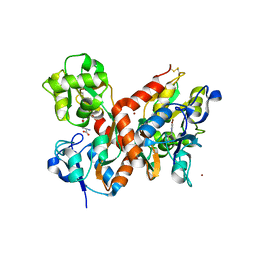 | | CRYSTAL STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) in complex with bromo-willardiine (Control for the crystal titration experiments) | | Descriptor: | 2-AMINO-3-(5-BROMO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2, ZINC ION | | Authors: | Jin, R, Gouaux, E. | | Deposit date: | 2002-10-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the Function, Conformational Plasticity, and Dimer-Dimer Contacts of the GluR2 Ligand-Binding Core: Studies of 5-Substituted Willardiines and GluR2 S1S2 in the Crystal
Biochemistry, 42, 2003
|
|
3PBA
 
 | | Crystal structure of PPARgamma ligand binding domain in complex with monosulfate tetrabromo-bisphenol A (MonoTBBPA) | | Descriptor: | 2,6-dibromo-4-[2-(3,5-dibromo-4-hydroxyphenyl)propan-2-yl]phenyl hydrogen sulfate, Peroxisome proliferator-activated receptor gamma, S-1,2-PROPANEDIOL | | Authors: | le Maire, A, Bourguet, W. | | Deposit date: | 2010-10-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of Novel Ligands of ER{alpha}, Er{beta}, and PPAR{gamma}: The Case of Halogenated Bisphenol A and Their Conjugated Metabolites.
Toxicol Sci, 122, 2011
|
|
7V8F
 
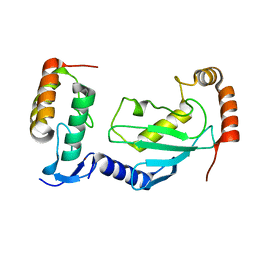 | | Crystal structure of UBE2L3 bound to HOIP RING1 domain. | | Descriptor: | E3 ubiquitin-protein ligase RNF31, Ubiquitin-conjugating enzyme E2 L3, ZINC ION | | Authors: | Liu, J, Wang, Y, Pan, L. | | Deposit date: | 2021-08-22 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Mechanistic insights into the subversion of the linear ubiquitin chain assembly complex by the E3 ligase IpaH1.4 of Shigella flexneri.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1NHB
 
 | |
1A52
 
 | |
2XNW
 
 | | XPT-PBUX C74U RIBOSWITCH FROM B. SUBTILIS BOUND TO A TRIAZOLO- TRIAZOLE-DIAMINE LIGAND IDENTIFIED BY VIRTUAL SCREENING | | Descriptor: | 3,6-diamino-1,5-dihydro[1,2,4]triazolo[4,3-b][1,2,4]triazol-4-ium, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Daldrop, P, Reyes, F.E, Robinson, D.A, Hammond, C.M, Lilley, D.M.J, Brenk, R. | | Deposit date: | 2010-08-06 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel ligands for a purine riboswitch discovered by RNA-ligand docking.
Chem. Biol., 18, 2011
|
|
9H9T
 
 | | Crystal structure of NEDD4 HECT domain in complex with covalent inhibitor | | Descriptor: | Isoform 4 of E3 ubiquitin-protein ligase NEDD4, ethyl 4-[4-(2-chloranyl-5,6-dihydrobenzo[b][1]benzazepin-11-yl)butylamino]butanoate | | Authors: | Cecatiello, V, Maspero, E, Polo, S, Pasqualato, S. | | Deposit date: | 2024-10-31 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure-based design of potent and selective inhibitors of the HECT ligase NEDD4.
Commun Chem, 8, 2025
|
|
9IQT
 
 | | structure of niacin-HCA2-Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Liu, Y, Zhou, Z. | | Deposit date: | 2024-07-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ligand Recognition and Activation Mechanism of the Alicarboxylic Acid Receptors.
J.Mol.Biol., 436, 2024
|
|
8Z30
 
 | | Crystal structure of HOIP PUB domain in complex with tolfenamic acid complex | | Descriptor: | 2-[(3-chloro-2-methylphenyl)amino]benzoic acid, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Zhong, F, Ruan, K. | | Deposit date: | 2024-04-14 | | Release date: | 2024-11-27 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Repurposing Tolfenamic Acid to Anchor the Uncharacterized Pocket of the PUB Domain for Proteolysis of the Atypical E3 Ligase HOIP.
Acs Chem.Biol., 19, 2024
|
|
8Z36
 
 | | Crystal structure of HOIP PUB domain in complex with sertraline complex | | Descriptor: | (1S,4S)-4-(3,4-dichlorophenyl)-N-methyl-1,2,3,4-tetrahydronaphthalen-1-amine, E3 ubiquitin-protein ligase RNF31 | | Authors: | Zhong, F, Ruan, K. | | Deposit date: | 2024-04-14 | | Release date: | 2024-11-27 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Repurposing Tolfenamic Acid to Anchor the Uncharacterized Pocket of the PUB Domain for Proteolysis of the Atypical E3 Ligase HOIP.
Acs Chem.Biol., 19, 2024
|
|
7U56
 
 | |
9H9O
 
 | |
6HCC
 
 | | STRUCTURE OF GLUA2 LIGAND-BINDING DOMAIN (S1S2J-N775S) IN COMPLEX WITH GLUTAMATE AND TDPAM02 AT 1.6 A RESOLUTION. | | Descriptor: | 6,6'-(ETHANE-1,2-DIYL)BIS(4-CYCLOPROPYL-3,4-DIHYDRO-2H-1,2,4-BENZOTHIADIAZINE 1,1-DIOXIDE), ACETATE ION, CHLORIDE ION, ... | | Authors: | Laulumaa, S, Hansen, K.V, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2018-08-14 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.617 Å) | | Cite: | Crystal Structures of Potent Dimeric Positive Allosteric Modulators at the Ligand-Binding Domain of the GluA2 Receptor.
Acs Med.Chem.Lett., 10, 2019
|
|
9EQH
 
 | | WWP2 WW2-2,3-linker-HECT (WWP2-LH) | | Descriptor: | GLYCEROL, Isoform 2 of NEDD4-like E3 ubiquitin-protein ligase WWP2, SODIUM ION | | Authors: | Dudey, A.P, Hemmings, A.M. | | Deposit date: | 2024-03-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Expanding the inhibitor space of the WWP1 and WWP2 HECT E3 ligases.
J Enzyme Inhib Med Chem, 39, 2024
|
|
1AQF
 
 | | PYRUVATE KINASE FROM RABBIT MUSCLE WITH MG, K, AND L-PHOSPHOLACTATE | | Descriptor: | L-PHOSPHOLACTATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Larsen, T.M, Benning, M.M, Wesenberg, G.E, Rayment, I, Reed, G.H. | | Deposit date: | 1997-07-29 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ligand-induced domain movement in pyruvate kinase: structure of the enzyme from rabbit muscle with Mg2+, K+, and L-phospholactate at 2.7 A resolution.
Arch.Biochem.Biophys., 345, 1997
|
|
