5MCK
 
 | | Radiation damage to GH7 Family Cellobiohydrolase from Daphnia pulex: Dose (DWD) 16.2 MGy | | Descriptor: | Cellobiohydrolase CHBI, GLYCEROL, SULFATE ION | | Authors: | Bury, C.S, McGeehan, J.E, Ebrahim, A, Garman, E.F. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | OH cleavage from tyrosine: debunking a myth.
J Synchrotron Radiat, 24, 2017
|
|
7A2J
 
 | |
7A2V
 
 | |
7A37
 
 | |
5MQY
 
 | | CATHEPSIN L IN COMPLEX WITH 4-[1,3-benzodioxol-5-ylmethyl(2-phenoxyethyl)amino]-5-fluoropyrimidine-2-carbonitrile | | Descriptor: | 1,2-ETHANEDIOL, 4-[1,3-benzodioxol-5-ylmethyl(2-phenoxyethyl)amino]-5-fluoropyrimidine-2-carbonitrile, Cathepsin L1 | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-12-21 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Prospective Evaluation of Free Energy Calculations for the Prioritization of Cathepsin L Inhibitors.
J. Med. Chem., 60, 2017
|
|
7A28
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-617 | | Descriptor: | 1-cycloheptyl-3-(4-methoxy-3-{4-[4-(1H-1,2,3,4-tetrazol-5-yl)phenoxy]butoxy}phenyl)-4,4-dimethyl-4,5-dihydro-1H-pyrazol-5-one, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2020-08-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-617
To be published
|
|
7A2Q
 
 | |
5MDR
 
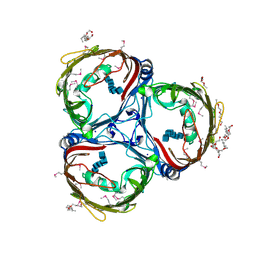 | | Crystal structure of in vitro folded Chitoporin VhChip from Vibrio harveyi in complex with chitohexaose | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitoporin, ... | | Authors: | Zahn, M, van den Berg, B. | | Deposit date: | 2016-11-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for chitin acquisition by marine Vibrio species.
Nat Commun, 9, 2018
|
|
5MR0
 
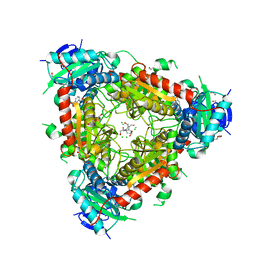 | | Thermophilic archaeal branched-chain amino acid transaminases from Geoglobus acetivorans and Archaeoglobus fulgidus: biochemical and structural characterisation | | Descriptor: | 1,2-ETHANEDIOL, 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, CHLORIDE ION, ... | | Authors: | Isupov, M.N, Littlechild, J.A, James, P, Sayer, C, Sutter, J.M, Schmidt, M, Schoenheit, P. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the ArchaeaGeoglobus acetivoransandArchaeoglobus fulgidus: Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
7A33
 
 | |
7A2W
 
 | |
7A3A
 
 | |
5MT6
 
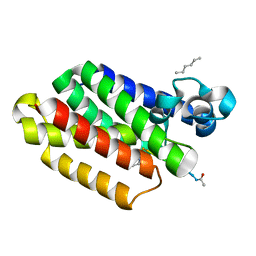 | |
7AAQ
 
 | | sugar/H+ symporter STP10 in outward occluded conformation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, ACETATE ION, ... | | Authors: | Bavnhoej, L, Paulsen, P.A, Pedersen, B.P. | | Deposit date: | 2020-09-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Molecular mechanism of sugar transport in plants unveiled by structures of glucose/H + symporter STP10.
Nat.Plants, 7, 2021
|
|
7A2P
 
 | |
5MTQ
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT511 | | Descriptor: | 2-[4-[(4-cyclohexyl-1,2,3-triazol-1-yl)methyl]-2-oxidanyl-phenoxy]benzenecarbonitrile, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
5MUB
 
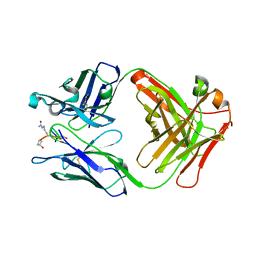 | | ACC1 Fab fragment in complex with citrullinated C1 epitope of CII (CG05) | | Descriptor: | ACC1 Fab fragment heavy chain, ACC1 Fab fragment light chain, triple-helical peptide containing the citrullinated C1 epitope of collagen type II,Collagen alpha-1(II) chain,triple-helical peptide containing the citrullinated C1 epitope of collagen type II | | Authors: | Dobritzsch, D, Holmdahl, R, Ge, C. | | Deposit date: | 2017-01-13 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Anti-citrullinated protein antibodies cause arthritis by cross-reactivity to joint cartilage.
JCI Insight, 2, 2017
|
|
5MGA
 
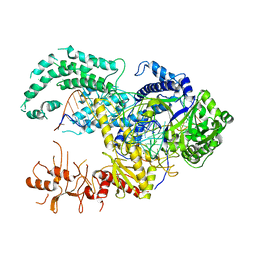 | |
5MVT
 
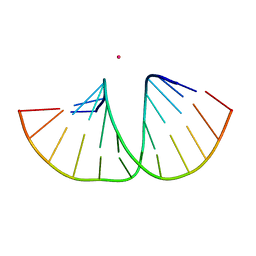 | | Crystal structure of an A-DNA dodecamer featuring an alternating pyrimidine-purine sequence | | Descriptor: | COBALT (III) ION, DNA | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7A2N
 
 | |
7A2M
 
 | |
6SCR
 
 | |
5MW9
 
 | | Complex between the Leucine Zipper (LZ) and Centrosomin-motif 2 (CM2) domains of Drosophila melanogaster Centrosomin (Cnn) - L535E mutant form | | Descriptor: | Centrosomin, ZINC ION | | Authors: | Feng, Z, Johnson, S, Raff, J.W, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Mitotic Centrosome Assembly in Flies.
Cell, 169, 2017
|
|
7A2R
 
 | |
5MWO
 
 | | Structure of Mycobacterium Tuberculosis Transcriptional Regulatory Repressor Protein (EthR) in complex with fragment 7E8. | | Descriptor: | (5-methyl-1-benzothiophen-2-yl)methanol, 1,2-ETHANEDIOL, HTH-type transcriptional regulator EthR | | Authors: | Mendes, V, Chan, D.S.-H, Thomas, S.E, McConnell, B, Matak-Vinkovic, D, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2017-01-18 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Fragment Screening against the EthR-DNA Interaction by Native Mass Spectrometry.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
