1OGA
 
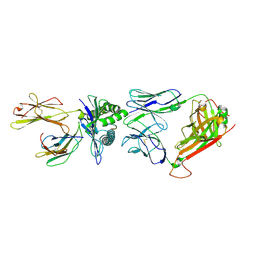 | | A structural basis for immunodominant human T-cell receptor recognition. | | Descriptor: | BETA-2-MICROGLOBULIN, GILGFVFTL, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Stewart-Jones, G.B.E, McMichael, A.J, Bell, J.I, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2003-04-28 | | Release date: | 2003-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Structural Basis for Immunodominant Human T Cell Receptor Recognition
Nat.Immunol., 4, 2003
|
|
5C5W
 
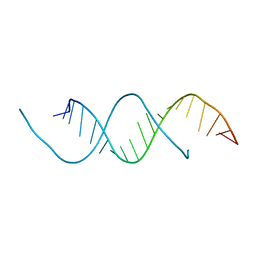 | | 1.25 A resolution structure of an RNA 20-mer | | Descriptor: | RNA (5'-R(P*CP*CP*UP*GP*AP*GP*UP*UP*CP*AP*AP*UP*UP*CP*UP*AP*GP*CP*G)-3') | | Authors: | Stewart, M, Valkov, E. | | Deposit date: | 2015-06-22 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 1.25 angstrom resolution structure of an RNA 20-mer that binds to the TREX2 complex.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
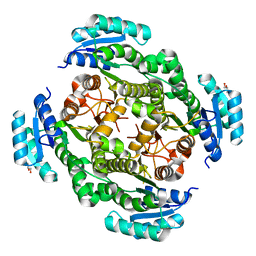
7QK1
 
 | |
7KRK
 
 | |
7QL0
 
 | |
4HIT
 
 | |
3EYC
 
 | | New crystal structure of human tear lipocalin in complex with 1,4-butanediol in space group P21 | | Descriptor: | 1,4-BUTANEDIOL, Lipocalin-1 | | Authors: | Breustedt, D.A, Keil, L, Skerra, A. | | Deposit date: | 2008-10-20 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A new crystal form of human tear lipocalin reveals high flexibility in the loop region and induced fit in the ligand cavity
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5FBB
 
 | | S1 nuclease from Aspergillus oryzae in complex with phosphate and adenosine 5'-monophosphate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Catalytic Properties of S1 Nuclease from Aspergillus oryzae Responsible for Substrate Recognition, Cleavage, Non-Specificity, and Inhibition.
PLoS ONE, 11, 2016
|
|
5BWK
 
 | | 6.0 A Crystal structure of a Get3-Get4-Get5 intermediate complex from S.cerevisiae | | Descriptor: | ATPase GET3, Golgi to ER traffic protein 4, Ubiquitin-like protein MDY2, ... | | Authors: | Gristick, H.B, Chartron, J.W, Clemons, W.M. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Mechanism of Assembly of a Substrate Transfer Complex during Tail-anchored Protein Targeting.
J.Biol.Chem., 290, 2015
|
|
8U6G
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 3-(2-(2-(3-acryloyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-1-yl)ethoxy)-4-chlorophenoxy)-5-chlorobenzonitrile (JLJ744), a non-nucleoside inhibitor | | Descriptor: | 3-chloro-5-{4-chloro-2-[2-(2-oxo-3-propanoyl-2,3-dihydro-1H-benzimidazol-1-yl)ethoxy]phenoxy}benzonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Prucha, G, Carter, Z, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
7QKI
 
 | |
5FBM
 
 | | Crystal Structure of Histone Like Protein (HLP) from Streptococcus mutans Refined to 1.9 A Resolution | | Descriptor: | DNA-binding protein HU | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, O'Neil, P, Biswas, I. | | Deposit date: | 2015-12-14 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of histone-like protein from Streptococcus mutans refined to 1.9 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
7QKM
 
 | |
2EM9
 
 | | Solution structure of the C2H2 type zinc finger (region 367-399) of human Zinc finger protein 224 | | Descriptor: | ZINC ION, Zinc finger protein 224 | | Authors: | Tomizawa, T, Tochio, N, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 367-399) of human Zinc finger protein 224
To be Published
|
|
7QL4
 
 | |
1OK4
 
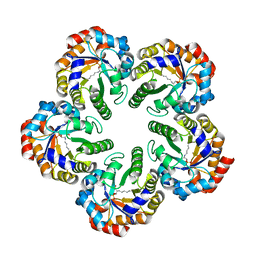 | | Archaeal fructose 1,6-bisphosphate aldolase covalently bound to the substrate dihydroxyacetone phosphate | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I | | Authors: | Lorentzen, E, Zwart, P, Stark, A, Hensel, R, Siebers, B, Pohl, E. | | Deposit date: | 2003-07-17 | | Release date: | 2003-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an archaeal class I aldolase and the evolution of (betaalpha)8 barrel proteins.
J. Biol. Chem., 278, 2003
|
|
2EMZ
 
 | | Solution structure of the C2H2 type zinc finger (region 628-660) of human Zinc finger protein 95 homolog | | Descriptor: | ZINC ION, Zinc finger protein 95 homolog | | Authors: | Tochio, N, Tomizawa, T, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 628-660) of human Zinc finger protein 95 homolog
To be Published
|
|
4MNL
 
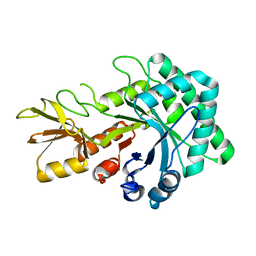 | | Crystal Structure of GH18 Chitinase (G77W/E119Q mutant) from Cycas revoluta in complex with (GlcNAc)4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Chitinase A | | Authors: | Umemoto, N, Numata, T, Ohnuma, T, Osawa, T, Taira, T, Fukamizo, T. | | Deposit date: | 2013-09-11 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of GH18 Chitinase form Cycad, Cycas revoluta
To be Published
|
|
7QKW
 
 | |
3EZQ
 
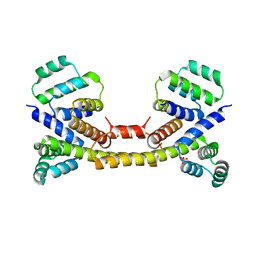 | | Crystal Structure of the Fas/FADD Death Domain Complex | | Descriptor: | Protein FADD, SODIUM ION, SULFATE ION, ... | | Authors: | Schwarzenbacher, R, Robinson, H, Stec, B, Riedl, S.J. | | Deposit date: | 2008-10-23 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The Fas-FADD death domain complex structure unravels signalling by receptor clustering
Nature, 457, 2009
|
|
1OK6
 
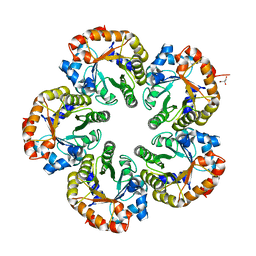 | | Orthorhombic crystal form of an Archaeal fructose 1,6-bisphosphate aldolase | | Descriptor: | FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I, GLYCEROL | | Authors: | Lorentzen, E, Zwart, P, Stark, A, Hensel, R, Siebers, B, Pohl, E. | | Deposit date: | 2003-07-18 | | Release date: | 2003-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an archaeal class I aldolase and the evolution of (betaalpha)8 barrel proteins.
J. Biol. Chem., 278, 2003
|
|
3FHD
 
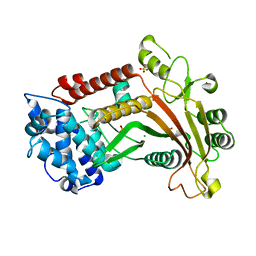 | | Crystal structure of the Shutoff and Exonuclease Protein from Kaposis Sarcoma Associated Herpesvirus | | Descriptor: | MAGNESIUM ION, ORF 37, SULFATE ION | | Authors: | Dahlroth, S.L, Gurmu, D, Schmitzberger, F, Haas, J, Erlandsen, H, Nordlund, P. | | Deposit date: | 2008-12-09 | | Release date: | 2009-11-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the shutoff and exonuclease protein from the oncogenic Kaposi's sarcoma-associated herpesvirus
Febs J., 276, 2009
|
|
7KLO
 
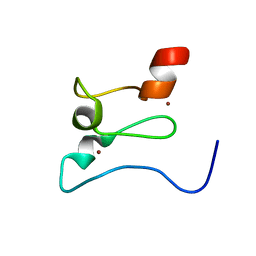 | |
4MMR
 
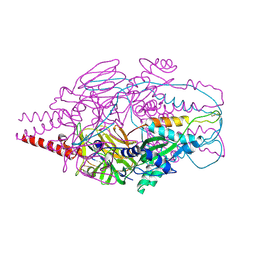 | | Crystal Structure of Prefusion-stabilized RSV F Variant Cav1 at pH 9.5 | | Descriptor: | Fusion glycoprotein F1 fused with Fibritin trimerization domain, Fusion glycoprotein F2 | | Authors: | Stewart-Jones, G.B.E, McLellan, J.S, Joyce, M.G, Sastry, M, Yang, Y, Graham, B.S, Kwong, P.D. | | Deposit date: | 2013-09-09 | | Release date: | 2013-11-20 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus.
Science, 342, 2013
|
|
7QGO
 
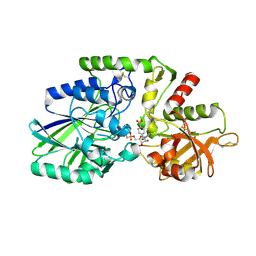 | | Human CD73 (ecto 5'-nucleotidase) in complex with MRS4602 (a 3-methyl-CMPCP derivative, compound 21 in paper) in the closed state (crystal form III) | | Descriptor: | 5'-nucleotidase, ZINC ION, [[(2~{R},3~{S},4~{R},5~{R})-5-[(4~{E})-4-[(4-methoxycarbonylphenyl)methoxyimino]-3-methyl-2-oxidanylidene-pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]methylphosphonic acid | | Authors: | Strater, N. | | Deposit date: | 2021-12-09 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structure-Activity Relationship of 3-Methylcytidine-5'-alpha , beta-methylenediphosphates as CD73 Inhibitors.
J.Med.Chem., 65, 2022
|
|
