8YS9
 
 | |
9EM4
 
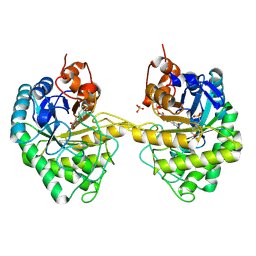 | | OPR3 variant Y364P in its dimeric form obtained with ammonium sulfate | | Descriptor: | 12-oxophytodienoate reductase 3, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Bijelic, A, Macheroux, P, Kerschbaumer, B. | | Deposit date: | 2024-03-07 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Analysis of homodimer formation in 12-oxophytodienoate reductase 3 in solutio and crystallo challenges the physiological role of the dimer.
Sci Rep, 14, 2024
|
|
8DYB
 
 | |
8YN7
 
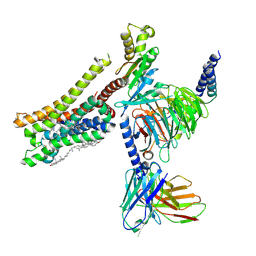 | | Cryo-EM structure of histamine H3 receptor in complex with immethridine and miniGo | | Descriptor: | 4-(1H-imidazol-4-ylmethyl)pyridine, Antibody fragment scFv16, CHOLESTEROL, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
9GD1
 
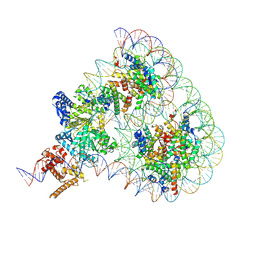 | | Structure of Chd1 bound to a hexasome-nucleosome complex with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9CUV
 
 | |
8G3U
 
 | | MBP-Mcl1 in complex with ligand 21 | | Descriptor: | (1'S,3aS,5R,16R,17S,19Z,21R,21aR)-6'-chloro-20-fluoro-21-{[(5S,9aS)-hexahydropyrazino[2,1-c][1,4]oxazin-8(1H)-yl]methyl}-21-methoxy-16,17-dimethyl-2,3,3',3a,4',16,17,18,21,21a-decahydro-2'H,6H,8H-15lambda~6~-spiro[10,12-(ethanediylidene)-15lambda~6~-furo[3,2-i][1,4]oxazepino[3,4-f][1,2,7]thiadiazacyclohexadecine-7,1'-naphthalene]-13,15,15(4H,14H)-trione, 1,2-ETHANEDIOL, Maltodextrin-binding protein, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
9DUV
 
 | |
6O8V
 
 | |
8G3S
 
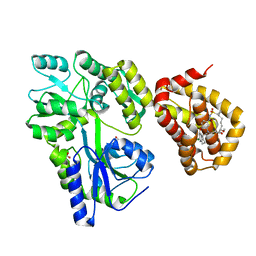 | | MBP-Mcl1 in complex with ligand 11 | | Descriptor: | (1'S,3aS,5R,16R,17S,19E,21S,21aR)-6'-chloro-21-methoxy-16,17-dimethyl-2,3,3',3a,4',16,17,18,21,21a-decahydro-2'H,6H,8H-15lambda~6~-spiro[10,12-etheno-15lambda~6~-furo[3,2-i][1,4]oxazepino[3,4-f][1,2,7]thiadiazacyclohexadecine-7,1'-naphthalene]-13,15,15(4H,14H)-trione, FORMIC ACID, Maltodextrin-binding protein, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
8DJF
 
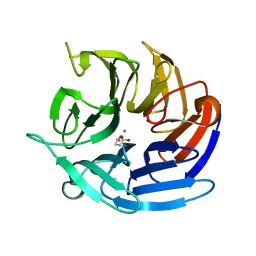 | | Crystal structure of RPA3624, a beta-propeller lactonase from Rhodopseudomonas palustris, with active-site bound tetrahedral intermediate | | Descriptor: | (5S)-5-methyloxolane-2,2-diol, CALCIUM ION, Gluconolactonase, ... | | Authors: | Bingman, C.A, Hall, B.W, Smith, R.W, Fox, B.G, Donohue, T.J. | | Deposit date: | 2022-06-30 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A broad specificity beta-propeller enzyme from Rhodopseudomonas palustris that hydrolyzes many lactones including gamma-valerolactone.
J.Biol.Chem., 299, 2022
|
|
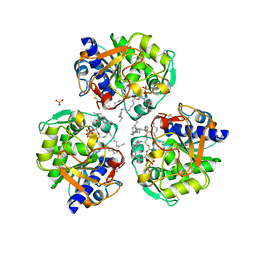
6OSZ
 
 | |
8G3X
 
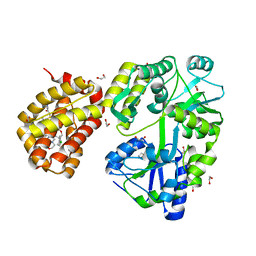 | | MBP-Mcl1 in complex with ligand 32 | | Descriptor: | 1,2-ETHANEDIOL, Maltodextrin-binding protein, Induced myeloid leukemia cell differentiation protein Mcl-1 chimera, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
8DKT
 
 | | Crystal Structure of Septin1 - Septin2 heterocomplex from Drosophila melanogaster | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | de Freitas, A.F, Leonardo, D.A, Cavini, I.A, Pereira, H.M, Garratt, R.C. | | Deposit date: | 2022-07-06 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Conservation and divergence of the G-interfaces of Drosophila melanogaster septins.
Cytoskeleton (Hoboken), 80, 2023
|
|
9CI8
 
 | | T cell receptor complex | | Descriptor: | T cell receptor delta constant, T cell receptor gamma constant 1, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Gully, B.S, Rossjohn, J. | | Deposit date: | 2024-07-02 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | T cell receptor complex
To Be Published
|
|
9EZ1
 
 | | Vitamin D receptor in complex with 1,4a,25-trihydroxyvitamin D3 | | Descriptor: | 1,4a,25-trihydroxyvitamin D3, ACETATE ION, Nuclear receptor coactivator 2, ... | | Authors: | Rochel, N. | | Deposit date: | 2024-04-10 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 4-Hydroxy-1 alpha ,25-Dihydroxyvitamin D 3 : Synthesis and Structure-Function Study.
Biomolecules, 14, 2024
|
|
9C0C
 
 | | E.coli GroEL apoenzyme | | Descriptor: | 60 kDa chaperonin | | Authors: | Watson, E.R, Lander, G.C. | | Deposit date: | 2024-05-25 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Bis-sulfonamido-2-phenylbenzoxazoles Validate the GroES/EL Chaperone System as a Viable Antibiotic Target.
J.Am.Chem.Soc., 146, 2024
|
|
9DN4
 
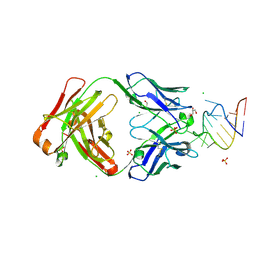 | | Crystal structure of a SARS-CoV-2 20-mer RNA in complex with FAB BL3-6S97N | | Descriptor: | CHLORIDE ION, FAB BL3-6S97N HEAVY CHAIN, FAB BL3-6S97N LIGHT CHAIN, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Hegde, S, Wang, J. | | Deposit date: | 2024-09-16 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a SARS-CoV-2 20-mer RNA in complex with FAB BL3-6S97N
To be published
|
|
8E2S
 
 | |
8Y2I
 
 | |
9CEZ
 
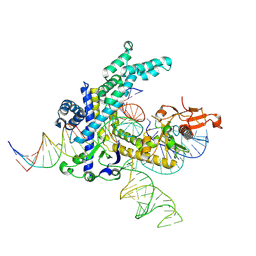 | | Spizellomyces punctatus Fanzor (SpuFz) State 6 | | Descriptor: | DNA (27-MER), DNA (5'-D(P*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9IY0
 
 | | anti-HEV mAb 8H3 | | Descriptor: | Heavy chain of 8H3 Fab, Light chain of 8H3 Fab, MAGNESIUM ION | | Authors: | Minghua, Z, Lizhi, Z, Yang, H, Ying, G, Shaowei, L. | | Deposit date: | 2024-07-29 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis for the synergetic neutralization of hepatitis E virus by antibody-antibody interaction
To Be Published
|
|
9CP0
 
 | |
9BGK
 
 | | Structure of V.cholera DdmDE (2D:1E) in complex with DNA | | Descriptor: | DdmE, Helicase/UvrB N-terminal domain-containing protein, MAGNESIUM ION, ... | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2024-04-19 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | DdmDE eliminates plasmid invasion by DNA-guided DNA targeting.
Cell, 187, 2024
|
|
9IYE
 
 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.39 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-07-30 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.39 A resolution.
To Be Published
|
|
