5FCZ
 
 | |
5E2O
 
 | |
5FE8
 
 | | Crystal structure of human PCAF bromodomain in complex with compound SL1126 (compound 12) | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Histone acetyltransferase KAT2B, ... | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
6ZPV
 
 | | Structure of Unliganded MgGH51 a-L-Arabinofuranosidase Crystal Type 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8ZJG
 
 | | Cryo-EM structure of human CMKLR1-Gi complex bound to chemerin | | Descriptor: | CHOLESTEROL, Chemerin-like receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, A.J, Liu, Y.Z, Ye, R.D. | | Deposit date: | 2024-05-14 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis for full-length chemerin recognition and signaling through chemerin receptor 1.
Commun Biol, 7, 2024
|
|
6G2O
 
 | | X-ray structure of NSD3-PWWP1 in complex with compound BI-9321 | | Descriptor: | Histone-lysine N-methyltransferase NSD3, [4-[5-(7-fluoranylquinolin-4-yl)-1-methyl-imidazol-4-yl]-3,5-dimethyl-phenyl]methanamine | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
5SXG
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B | | Descriptor: | 1,3-PROPANDIOL, DNA dC->dU-editing enzyme APOBEC-3B, IMIDAZOLE, ... | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2016-08-09 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Conformational Switch Regulates the DNA Cytosine Deaminase Activity of Human APOBEC3B.
Sci Rep, 7, 2017
|
|
2XO0
 
 | | xpt-pbuX C74U Riboswitch from B. subtilis bound to 24-diamino-1,3,5- triazine identified by virtual screening | | Descriptor: | 1,3,5-TRIAZINE-2,4-DIAMINE, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Daldrop, P, Reyes, F.E, Robinson, D.A, Hammond, C.M, Lilley, D.M.J, Batey, R.T, Brenk, R. | | Deposit date: | 2010-08-09 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel ligands for a purine riboswitch discovered by RNA-ligand docking.
Chem. Biol., 18, 2011
|
|
8Z6R
 
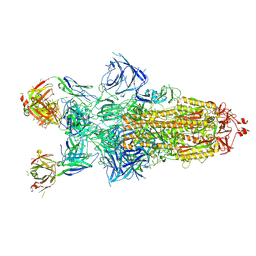 | | Structure of XBB.1.16 S trimer with 3 down-RBDs complex with antibody CYFN1006-1. | | Descriptor: | CYFN1006-1 heavy chain, CYFN1006-1 light chain, Spike glycoprotein,Fibritin,Spike glycoprotein,Fibritin,Spike glycoprotein,Fibritin,Expression Tag | | Authors: | Wang, Y.J, Sun, L. | | Deposit date: | 2024-04-19 | | Release date: | 2025-01-29 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structure of XBB.1.16 S trimer with 3 down-RBDs complex with antibody CYFN1006-1.
To Be Published
|
|
9M73
 
 | | Crystal structure of MBP-fused BIL1/BZR1 (21-104) in complex with double-stranded DNA contaning CACATATGTG | | Descriptor: | 1,2-ETHANEDIOL, E-box(CATATG)-containing DNA, Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Shohei, N, Masaru, T, Takuya, M. | | Deposit date: | 2025-03-09 | | Release date: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Single cis-elements in brassinosteroid-induced upregulated genes are insufficient to recruit both redox states of the BIL1/BZR1 DNA-binding domain.
Febs Lett., 2025
|
|
9C9O
 
 | | M. tuberculosis PKS13 acyltransferase (AT) domain in complex with SuFEx inhibitor CMX410 - reaction product | | Descriptor: | 4-({2,6-difluoro-4-[3-(methanesulfonamido)-1H-1,2,4-triazol-1-yl]phenyl}methoxy)phenyl hydrogen sulfate, PENTAETHYLENE GLYCOL, Polyketide synthase Pks13, ... | | Authors: | Krieger, I.V, Tang, S, Sacchetini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2024-06-14 | | Release date: | 2025-05-07 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | SuFEx-based antitubercular compound irreversibly inhibits Pks13.
Nature, 645, 2025
|
|
3JZ6
 
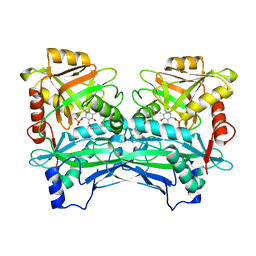 | | Crystal structure of Mycobacterium smegmatis Branched Chain Aminotransferase in complex with pyridoxal-5'-phosphate at 1.9 angstrom. | | Descriptor: | Branched-chain amino acid aminotransferase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Castell, A, Mille, C, Unge, T. | | Deposit date: | 2009-09-23 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of mycobacterial branched-chain aminotransferase: implications for inhibitor design.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5DCB
 
 | |
7Z8D
 
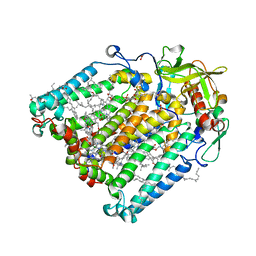 | | Structure of Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV by fixed-target serial synchrotron crystallography (room temperature, 26keV) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, ... | | Authors: | Gabdulkhakov, A.G, Selikhanov, G.K, Guenther, S, Petrova, T, Meents, A, Fufina, T.Y, Vasilieva, L.G. | | Deposit date: | 2022-03-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure of Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV by fixed-target serial synchrotron crystallography (room temperature, 26keV)
To Be Published
|
|
1KLJ
 
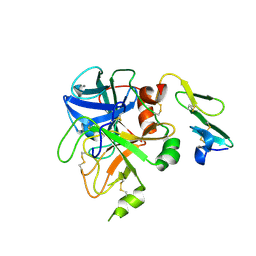 | | Crystal structure of uninhibited factor VIIa | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, factor VIIa | | Authors: | Sichler, K, Banner, D, D'Arcy, A, Hopfner, K.P, Huber, R, Bode, W, Kresse, G.B, Kopetzki, E, Brandstetter, H. | | Deposit date: | 2001-12-12 | | Release date: | 2002-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structures of uninhibited factor VIIa link its cofactor and substrate-assisted activation to specific interactions.
J.Mol.Biol., 322, 2002
|
|
1FMI
 
 | | CRYSTAL STRUCTURE OF HUMAN CLASS I ALPHA1,2-MANNOSIDASE | | Descriptor: | CALCIUM ION, ENDOPLASMIC RETICULUM ALPHA-MANNOSIDASE I, SULFATE ION | | Authors: | Vallee, F, Karaveg, K, Herscovics, A, Moremen, K.W, Howell, P.L. | | Deposit date: | 2000-08-17 | | Release date: | 2001-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for catalysis and inhibition of N-glycan processing class I alpha 1,2-mannosidases.
J.Biol.Chem., 275, 2000
|
|
5T2E
 
 | |
5HGB
 
 | | HLA*A2402 complexed with HIV nef138 8mer epitope | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2016-01-08 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Effects of a Single Escape Mutation on T Cell and HIV-1 Co-adaptation.
Cell Rep, 15, 2016
|
|
8TRV
 
 | | Structure of the EphA2 LBDCRD bound to FabS1C_C1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ephrin type-A receptor 2, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2024-07-17 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Synthetic antibodies targeting EphA2 induce diverse signaling-competent clusters with differential activation.
Protein Sci., 34, 2025
|
|
7ZD0
 
 | | Crystal structure of Pseudomonas aeruginosa S-adenosyl-L-homocysteine hydrolase inhibited by Cd2+ ions | | Descriptor: | 1,3-PROPANDIOL, 1,4-BUTANEDIOL, ADENOSINE, ... | | Authors: | Malecki, P.H, Gawel, M, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | A closer look at molecular mechanisms underlying inhibition of S -adenosyl-L-homocysteine hydrolase by transition metal cations.
Chem.Commun.(Camb.), 60, 2024
|
|
6BQY
 
 | |
5CT5
 
 | | Wild-type Bacillus subtilis lipase A with 10% [BMIM][Cl] | | Descriptor: | 1-butyl-3-methyl-1H-imidazol-3-ium, CHLORIDE ION, SULFATE ION, ... | | Authors: | Nordwald, E.M, Plaks, J.G, Snell, J.R, Sousa, M.C, Kaar, J.L. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Crystallographic Investigation of Imidazolium Ionic Liquid Effects on Enzyme Structure.
Chembiochem, 16, 2015
|
|
1AMT
 
 | |
5H9V
 
 | | Crystal structure of Regnase PIN domain, form I | | Descriptor: | Ribonuclease ZC3H12A, SODIUM ION | | Authors: | Yokogawa, M, Tsushima, T, Adachi, W, Noda, N.N, Inagaki, F. | | Deposit date: | 2015-12-29 | | Release date: | 2016-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
6BSK
 
 | | Human PIM1 kinase in complex with compound 12b | | Descriptor: | 1,2-ETHANEDIOL, 4-{6-[6-(propan-2-ylamino)-1H-indazol-1-yl]pyrazin-2-yl}benzoic acid, SULFATE ION, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2017-12-03 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.573 Å) | | Cite: | Discovery of 2,6-disubstituted pyrazine derivatives as inhibitors of CK2 and PIM kinases.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
