3KFO
 
 | | Crystal structure of the C-terminal domain from the nuclear pore complex component NUP133 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Nucleoporin NUP133 | | Authors: | Sampathkumar, P, Bonanno, J.B, Miller, S, Bain, K, Dickey, M, Gheyi, T, Almo, S.C, Rout, M, Sali, A, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-27 | | Release date: | 2010-01-26 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal domain of Saccharomyces cerevisiae Nup133, a component of the nuclear pore complex.
Proteins, 79, 2011
|
|
1DNC
 
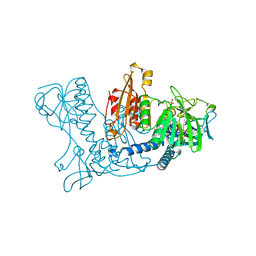 | | HUMAN GLUTATHIONE REDUCTASE MODIFIED BY DIGLUTATHIONE-DINITROSO-IRON | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, GLUTATHIONE REDUCTASE, ... | | Authors: | Becker, K, Savvides, S.N, Keese, M, Schirmer, R.H, Karplus, P.A. | | Deposit date: | 1998-02-20 | | Release date: | 1998-05-27 | | Last modified: | 2011-12-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enzyme inactivation through sulfhydryl oxidation by physiologic NO-carriers.
Nat.Struct.Biol., 5, 1998
|
|
1IHI
 
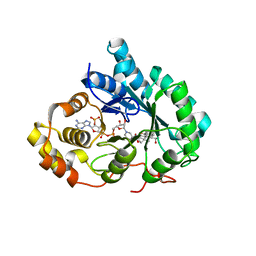 | | Crystal Structure of Human Type III 3-alpha-Hydroxysteroid Dehydrogenase/Bile Acid Binding Protein (AKR1C2) Complexed with NADP+ and Ursodeoxycholate | | Descriptor: | 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE, ISO-URSODEOXYCHOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jin, Y, Stayrook, S.E, Albert, R.H, Palackal, N.T, Penning, T.M, Lewis, M. | | Deposit date: | 2001-04-19 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human type III 3alpha-hydroxysteroid dehydrogenase/bile acid binding protein complexed with NADP(+) and ursodeoxycholate.
Biochemistry, 40, 2001
|
|
7V18
 
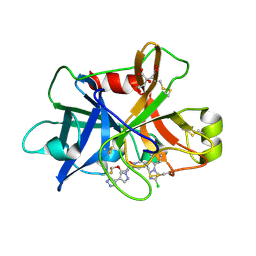 | | Factor XIa in Complex with Compound 3f | | Descriptor: | 2-[(1~{R})-3-[bis(fluoranyl)methoxy]-1-[4-(3-methyl-1,2,3-triazol-4-yl)pyrazol-1-yl]propyl]-5-[3-chloranyl-6-(4-chloranyl-1,2,3-triazol-1-yl)-2-fluoranyl-phenyl]-1-oxidanyl-pyridine, CITRIC ACID, Coagulation factor XIa light chain | | Authors: | Shaffer, P.L, Milligan, C.M. | | Deposit date: | 2022-05-11 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | Discovery of Potent and Orally Bioavailable Pyridine N-Oxide-Based Factor XIa Inhibitors through Exploiting Nonclassical Interactions.
J.Med.Chem., 65, 2022
|
|
1IHX
 
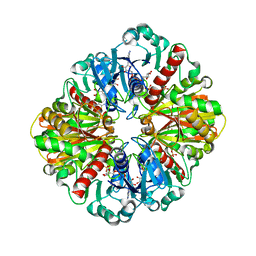 | | Crystal structure of two D-glyceraldehyde-3-phosphate dehydrogenase complexes: a case of asymmetry | | Descriptor: | GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE, SULFATE ION, THIONICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Shen, Y.-Q, Song, S.-Y, Lin, Z.-J. | | Deposit date: | 2001-04-20 | | Release date: | 2002-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of D-glyceraldehyde-3-phosphate dehydrogenase complexed with coenzyme analogues.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3V1W
 
 | | Molecular Basis for Multiple Ligand Binding of Calsequestrin and Potential Inhibition by Caffeine and Gallocatecin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Subramanian, A.K, Nissen, M.N, Lewis, K.M, Sanchez, E.J, Muralidharan, A.K, Kang, C. | | Deposit date: | 2011-12-10 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | Molecular Basis for Multiple Ligand Binding of Calsequestrin and Potential Inhibition by Caffeine and Gallocatecin
To be Published
|
|
6I3L
 
 | | Bilirubin oxidase from Myrothecium verrucaria, mutant W396F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bilirubin oxidase, COPPER (II) ION, ... | | Authors: | Koval, T, Svecova, L, Skalova, T, Kolenko, P, Duskova, J, Ostergaard, L.H, Dohnalek, J. | | Deposit date: | 2018-11-06 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Trp-His covalent adduct in bilirubin oxidase is crucial for effective bilirubin binding but has a minor role in electron transfer.
Sci Rep, 9, 2019
|
|
1DO8
 
 | | CRYSTAL STRUCTURE OF A CLOSED FORM OF HUMAN MITOCHONDRIAL NAD(P)+-DEPENDENT MALIC ENZYME | | Descriptor: | MALIC ENZYME, MANGANESE (II) ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yang, Z, Floyd, D.L, Loeber, G, Tong, L. | | Deposit date: | 1999-12-19 | | Release date: | 2000-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a closed form of human malic enzyme and implications for catalytic mechanism.
Nat.Struct.Biol., 7, 2000
|
|
1IM5
 
 | |
1IP7
 
 | | G129A HUMAN LYSOZYME | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
1ZNE
 
 | | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex | | Descriptor: | CADMIUM ION, HEXAN-1-OL, Major Urinary Protein | | Authors: | Malham, R, Johnstone, S, Bingham, R.J, Barratt, E, Phillips, S.E, Laughton, C.A, Homans, S.W. | | Deposit date: | 2005-05-11 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex.
J.Am.Chem.Soc., 127, 2005
|
|
6EBC
 
 | | OhrB (Organic Hydroperoxide Resistance protein) wild type from Chromobacterium violaceum and reduced by DTT | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, Organic hydroperoxide resistance protein | | Authors: | Domingos, R.M, Teixeira, R.D, Alegria, T.G.P, Vieira, P.S, Murakami, M.T, Netto, L.E.S. | | Deposit date: | 2018-08-06 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Substrate and product-assisted catalysis: molecular aspects behind structural switches along Organic Hydroperoxide Resistance Protein catalytic cycle
Acs Catalysis, 2020
|
|
1ZNL
 
 | | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex | | Descriptor: | CADMIUM ION, DECAN-1-OL, Major Urinary Protein | | Authors: | Malham, R, Johnstone, S, Bingham, R.J, Barratt, E, Phillips, S.E, Laughton, C.A, Homans, S.W. | | Deposit date: | 2005-05-11 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex.
J.Am.Chem.Soc., 127, 2005
|
|
5AAF
 
 | | Aurora A kinase bound to an imidazopyridine inhibitor (14a) | | Descriptor: | 3-((4-(6-chloro-2-(1,3-dimethyl-1H-pyrazol-4-yl)-3H-imidazo[4,5-b]pyridin-7-yl)-1H-pyrazol-1-yl)methyl)-N,N-dimethylbenzamide, AURORA KINASE A | | Authors: | McIntyre, P.J, Bayliss, R. | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | 7-(Pyrazol-4-Yl)-3H-Imidazo[4,5-B]Pyridine-Based Derivatives for Kinase Inhibition: Co-Crystallisation Studies with Aurora-A Reveal Distinct Differences in the Orientation of the Pyrazole N1-Substituent.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1INU
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1IO5
 
 | | HYDROGEN AND HYDRATION OF HEN EGG-WHITE LYSOZYME DETERMINED BY NEUTRON DIFFRACTION | | Descriptor: | LYSOZYME C | | Authors: | Niimura, N, Minezaki, Y, Nonaka, T, Castagna, J.C, Cipriani, F, Hoeghoej, P, Lehmann, M.S, Wilkinson, C. | | Deposit date: | 2001-01-14 | | Release date: | 2001-02-07 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | Neutron Laue diffractometry with an imaging plate provides an effective data collection regime for neutron protein crystallography.
Nat.Struct.Biol., 4, 1997
|
|
1Q3W
 
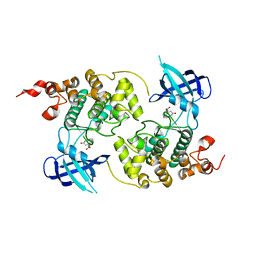 | | GSK-3 Beta complexed with Alsterpaullone | | Descriptor: | 9-NITRO-5,12-DIHYDRO-7H-BENZO[2,3]AZEPINO[4,5-B]INDOL-6-ONE, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Bertrand, J.A, Thieffine, S, Vulpetti, A, Cristiani, C, Valsasina, B, Knapp, S, Kalisz, H.M, Flocco, M. | | Deposit date: | 2003-08-01 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of the Gsk-3Beta Active Site Using Selective and Non-selective ATP-Mimetic Inhibitors
J.Mol.Biol., 333, 2003
|
|
1IR7
 
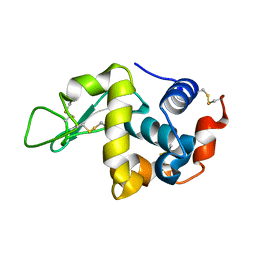 | | IM mutant of lysozyme | | Descriptor: | lysozyme | | Authors: | Ohmura, T, Ueda, T, Hashimoto, Y, Imoto, T. | | Deposit date: | 2001-09-19 | | Release date: | 2001-10-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tolerance of point substitution of methionine for isoleucine in hen egg white lysozyme.
Protein Eng., 14, 2001
|
|
5WJQ
 
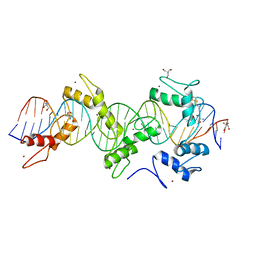 | | mouseZFP568-ZnF2-11 in complex with DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (28-MER), ZINC ION, ... | | Authors: | Patel, A, Cheng, X. | | Deposit date: | 2017-07-24 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | DNA Conformation Induces Adaptable Binding by Tandem Zinc Finger Proteins.
Cell, 173, 2018
|
|
1IP5
 
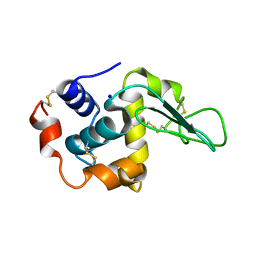 | | G105A HUMAN LYSOZYME | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
1IR9
 
 | | IM mutant of lysozyme | | Descriptor: | lysozyme | | Authors: | Ohmura, T, Ueda, T, Hashimoto, Y, Imoto, T. | | Deposit date: | 2001-09-19 | | Release date: | 2001-10-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tolerance of point substitution of methionine for isoleucine in hen egg white lysozyme.
Protein Eng., 14, 2001
|
|
5ABX
 
 | |
6IFI
 
 | |
7XA9
 
 | | Structure of Arabidopsis thaliana CLCa | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chloride channel protein CLC-a, MAGNESIUM ION, ... | | Authors: | Ji, S, Jin, H, Kaiming, Z, Mingxing, W, Shanshan, L, Long, C. | | Deposit date: | 2022-03-17 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of the plant nitrate transporter AtCLCa reveals characteristics of the anion-binding site and the ATP-binding pocket.
J.Biol.Chem., 299, 2023
|
|
3US3
 
 | | Recombinant rabbit skeletal calsequestrin-MPD complex | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sanchez, E.J, Lewis, K.M, Nissen, M.S, Munske, G.R, Kang, C. | | Deposit date: | 2011-11-22 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | Glycosylation of skeletal calsequestrin: implications for its function.
J.Biol.Chem., 287, 2012
|
|
