8U6N
 
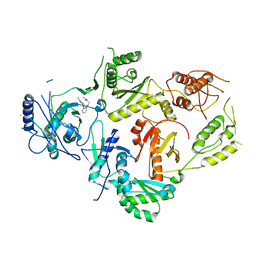 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 3-(2-((6-cyanonaphthalen-1-yl)oxy)phenoxy)-N,N-dimethylpropanamide (JLJ752), a non-nucleoside inhibitor | | Descriptor: | 3-{2-[(6-cyanonaphthalen-1-yl)oxy]phenoxy}-N,N-dimethylpropanamide, Gag-Pol polyprotein, p51 RT | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6I
 
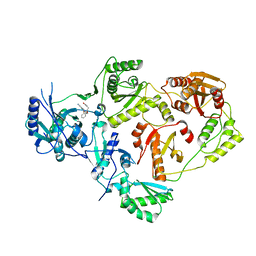 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(2-(2-((2-cyanoindolizin-8-yl)oxy)phenoxy)ethyl)-N-methylacrylamide (JLJ745), a non-nucleoside inhibitor | | Descriptor: | MAGNESIUM ION, N-[2-(2-{[(4R)-2-cyanoindolizin-8-yl]oxy}phenoxy)ethyl]-N-methylpropanamide, Reverse transcriptase/ribonuclease H, ... | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6L
 
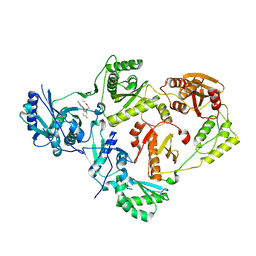 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(2-(5-chloro-2-((6-cyanonaphthalen-1-yl)oxy)phenoxy)ethyl)-N-methylacrylamide (JLJ748), a non-nucleoside inhibitor | | Descriptor: | MAGNESIUM ION, N-(2-{5-chloro-2-[(6-cyanonaphthalen-1-yl)oxy]phenoxy}ethyl)-N-methylprop-2-enamide, Reverse transcriptase/ribonuclease H, ... | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6J
 
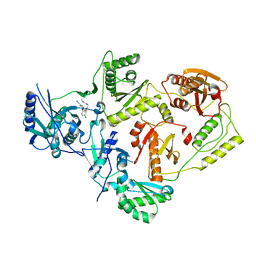 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(2-(5-chloro-2-((2-cyanoindolizin-8-yl)oxy)phenoxy)ethyl)-N-methylacrylamide (JLJ746), a non-nucleoside inhibitor | | Descriptor: | MAGNESIUM ION, N-[2-(5-chloro-2-{[(4R)-2-cyanoindolizin-8-yl]oxy}phenoxy)ethyl]-N-methylpropanamide, Reverse transcriptase/ribonuclease H, ... | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6F
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(2-(5-chloro-2-(3-chloro-5-cyanophenoxy)phenoxy)ethyl)-N-methylacrylamide (JLJ742), a non-nucleoside inhibitor | | Descriptor: | N-{2-[5-chloro-2-(3-chloro-5-cyanophenoxy)phenoxy]ethyl}-N-methylprop-2-enamide, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Prucha, G, Carter, Z, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6M
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(2-(2-((6-chloro-2-cyanoindolizin-8-yl)oxy)phenoxy)ethyl)-N-methylacrylamide (JLJ751), a non-nucleoside inhibitor | | Descriptor: | N-[2-(2-{[(4R)-6-chloro-2-cyanoindolizin-8-yl]oxy}phenoxy)ethyl]-N-methylpropanamide, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6T
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-(2-(2-(3-acryloyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-1-yl)ethoxy)phenoxy)-2-naphthonitrile (JLJ758), a non-nucleoside inhibitor | | Descriptor: | 5-(2-{2-[2-oxo-3-(prop-2-enoyl)-2,3-dihydro-1H-benzimidazol-1-yl]ethoxy}phenoxy)naphthalene-2-carbonitrile, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6D
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(2-(4-chloro-3-(3-chloro-5-cyanophenoxy)phenoxy)ethyl)-N-methylacrylamide (JLJ736), a non-nucleoside inhibitor | | Descriptor: | N-{2-[4-chloro-3-(3-chloro-5-cyanophenoxy)phenoxy]ethyl}-N-methylprop-2-enamide, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Hollander, K, Carter, Z, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6K
 
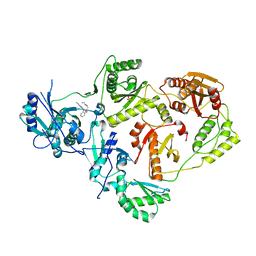 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(2-(2-((6-cyanonaphthalen-1-yl)oxy)phenoxy)ethyl)-N-methylacrylamide (JLJ747), a non-nucleoside inhibitor | | Descriptor: | MAGNESIUM ION, N-(2-{2-[(6-cyanonaphthalen-1-yl)oxy]phenoxy}ethyl)-N-methylprop-2-enamide, PHOSPHATE ION, ... | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6S
 
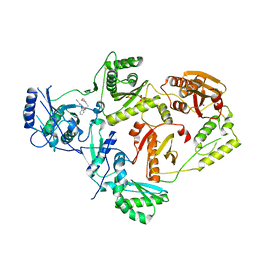 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 8-(2-(3-morpholino-3-oxopropoxy)phenoxy)indolizine-2-carbonitrile (JLJ757), a non-nucleoside inhibitor | | Descriptor: | (4S)-8-{2-[3-(morpholin-4-yl)-3-oxopropoxy]phenoxy}indolizine-2-carbonitrile, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6C
 
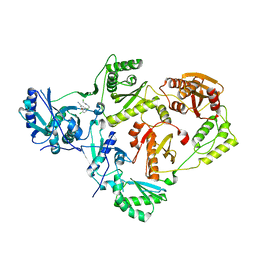 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 2-chloro-N-(4-chloro-3-(3-chloro-5-cyanophenoxy)phenethyl)acetamide (JLJ732), a non-nucleoside inhibitor | | Descriptor: | 2-chloro-N-{2-[4-chloro-3-(3-chloro-5-cyanophenoxy)phenyl]ethyl}acetamide, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Hollander, K, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6H
 
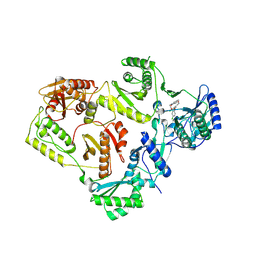 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 3-(2-(2-(3-acryloyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-1-yl)ethoxy)-4-chlorophenoxy)-5-chlorobenzonitrile (JLJ744), a non-nucleoside inhibitor | | Descriptor: | 3-chloro-5-{4-chloro-2-[2-(2-oxo-3-propanoyl-2,3-dihydro-1H-benzimidazol-1-yl)ethoxy]phenoxy}benzonitrile, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Prucha, G, Carter, Z, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6R
 
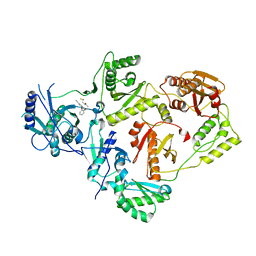 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 3-(2-((2-cyanoindolizin-8-yl)oxy)phenoxy)-N-(2,2-difluoroethyl)propanamide (JLJ756), a non-nucleoside inhibitor | | Descriptor: | 3-(2-{[(4R)-2-cyanoindolizin-8-yl]oxy}phenoxy)-N-(2,2-difluoroethyl)propanamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8UCD
 
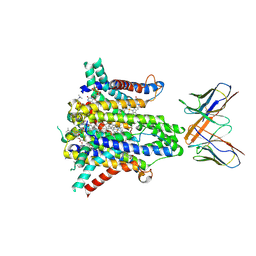 | | Cryo-EM structure of human STEAP1 in complex with AMG 509 Fab | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, AMG 509 anti-STEAP1 Fab, heavy chain, ... | | Authors: | Li, F, Bailis, J.M, Zhang, H. | | Deposit date: | 2023-09-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | AMG 509 (Xaluritamig), an Anti-STEAP1 XmAb 2+1 T-cell Redirecting Immune Therapy with Avidity-Dependent Activity against Prostate Cancer.
Cancer Discov, 14, 2024
|
|
7DTE
 
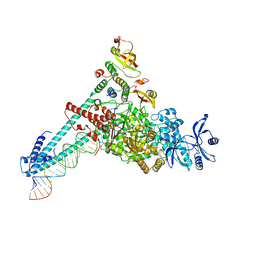 | | SARS-CoV-2 RdRP catalytic complex with T33-1 RNA | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (33-MER), ... | | Authors: | Wang, Q, Gong, P. | | Deposit date: | 2021-01-04 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Remdesivir overcomes the S861 roadblock in SARS-CoV-2 polymerase elongation complex.
Cell Rep, 37, 2021
|
|
5GUH
 
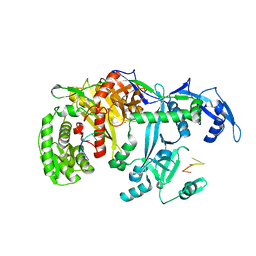 | | Crystal structure of silkworm PIWI-clade Argonaute Siwi bound to piRNA | | Descriptor: | MAGNESIUM ION, PIWI, RNA (28-MER) | | Authors: | Matsumoto, N, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Silkworm PIWI-Clade Argonaute Siwi Bound to piRNA
Cell, 167, 2016
|
|
5GIT
 
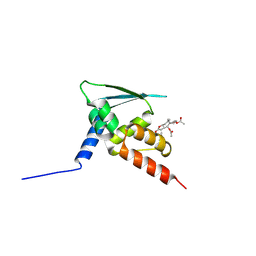 | | BTB domain of KEAP1 in complex with XX3 | | Descriptor: | Kelch-like ECH-associated protein 1, [(3aS,5R,5aS,6S,8S,8aS,9S,9aR)-9-acetyloxy-8-hydroxy-5,8a-dimethyl-1-methylidene-2-oxo-4,5,5a,6,7,8,9,9a-octahydro-3aH-azuleno[6,5-b]furan-6-yl] acetate | | Authors: | Zhu, L.L, Li, H.L, Wu, F.S, Xiong, R. | | Deposit date: | 2016-06-25 | | Release date: | 2017-06-28 | | Last modified: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Britanin Ameliorates Cerebral Ischemia-Reperfusion Injury by Inducing the Nrf2 Protective Pathway.
Antioxid. Redox Signal., 27, 2017
|
|
5G3O
 
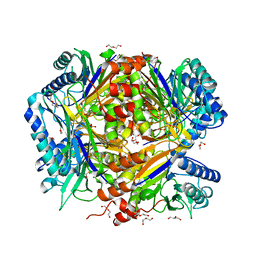 | | Bacillus cereus formamidase (BceAmiF) inhibited with urea. | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, DI(HYDROXYETHYL)ETHER, FORMAMIDASE, ... | | Authors: | Gavira, J.A, Martinez-Rodriguez, S, Conejero-Muriel, M. | | Deposit date: | 2016-04-29 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A novel cysteine carbamoyl-switch is responsible for the inhibition of formamidase, a nitrilase superfamily member.
Arch.Biochem.Biophys., 662, 2019
|
|
8USS
 
 | | IL17A complexed to Compound 7 | | Descriptor: | 4,5-dichloro-N-[(1S)-1-cyclohexyl-2-{[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]amino}-2-oxoethyl]-1H-pyrrole-2-carboxamide, CHLORIDE ION, Interleukin-17A | | Authors: | Argiriadi, M.A, Ramos, A.L. | | Deposit date: | 2023-10-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of Small Molecule Interleukin 17A Inhibitors with Novel Binding Mode and Stoichiometry: Optimization of DNA-Encoded Chemical Library Hits to In Vivo Active Compounds.
J.Med.Chem., 67, 2024
|
|
8USR
 
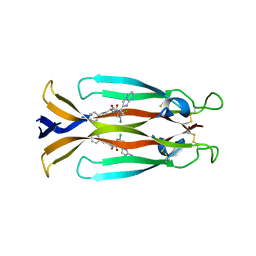 | | IL17A homodimer complexed to Compound 23 | | Descriptor: | Interleukin-17A, ~{N}-[(2~{S})-1-[[(1~{S})-1-(8~{a}~{H}-imidazo[1,2-a]pyrimidin-2-yl)ethyl]amino]-1-oxidanylidene-4-phenyl-butan-2-yl]-4,5-bis(chloranyl)-1~{H}-pyrrole-2-carboxamide | | Authors: | Argiriadi, M.A, Ramos, A.L. | | Deposit date: | 2023-10-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of Small Molecule Interleukin 17A Inhibitors with Novel Binding Mode and Stoichiometry: Optimization of DNA-Encoded Chemical Library Hits to In Vivo Active Compounds.
J.Med.Chem., 67, 2024
|
|
6M2N
 
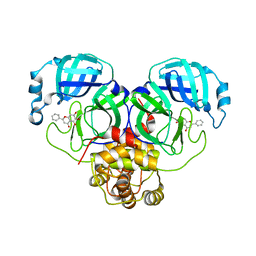 | | SARS-CoV-2 3CL protease (3CL pro) in complex with a novel inhibitor | | Descriptor: | 3C-like proteinase, 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one | | Authors: | Su, H.X, Zhao, W.F, Li, M.J, Xie, H, Xu, Y.C. | | Deposit date: | 2020-02-28 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients.
Acta Pharmacol.Sin., 41, 2020
|
|
6M2Q
 
 | | SARS-CoV-2 3CL protease (3CL pro) apo structure (space group C21) | | Descriptor: | 3C-like proteinase | | Authors: | Su, H.X, Zhao, W.F, Li, M.J, Xie, H, Xu, Y.C. | | Deposit date: | 2020-02-28 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients.
Acta Pharmacol.Sin., 41, 2020
|
|
7END
 
 | |
7EN9
 
 | | Crystal structure of SARS-CoV-2 3CLpro in complex with the non-covalent inhibitor WU-02 | | Descriptor: | 3C-like proteinase, 5-bromanyl-~{N}-methyl-3-nitro-2-[(4~{R},5~{S})-2-(7-oxidanylisoquinolin-4-yl)carbonyl-4-phenyl-2,7-diazaspiro[4.4]nonan-7-yl]benzamide | | Authors: | Hou, N, Peng, C, Hu, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 3CLpro.
Acs Cent.Sci., 9, 2023
|
|
7EN8
 
 | | Crystal structure of SARS-CoV-2 3CLpro in complex with the non-covalent inhibitor WU-04 | | Descriptor: | 3C-like proteinase, GLYCEROL, ~{N}-[(1~{S},2~{R})-2-[[4-bromanyl-2-(methylcarbamoyl)-6-nitro-phenyl]amino]cyclohexyl]isoquinoline-4-carboxamide | | Authors: | Hou, N, Peng, C, Hu, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Development of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 3CLpro.
Acs Cent.Sci., 9, 2023
|
|
