2ZIH
 
 | |
4HU9
 
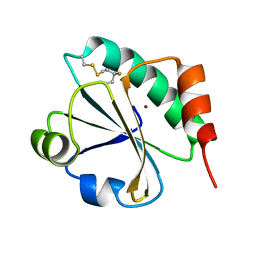 | | E. coli thioredoxin variant with (4S)-FluoroPro76 as single proline residue | | Descriptor: | COPPER (II) ION, Thioredoxin-1 | | Authors: | Scharer, M.A, Rubini, M, Capitani, G, Glockshuber, R. | | Deposit date: | 2012-11-02 | | Release date: | 2013-05-29 | | Last modified: | 2017-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | (4R)- and (4S)-Fluoroproline in the Conserved cis-Prolyl Peptide Bond of the Thioredoxin Fold: Tertiary Structure Context Dictates Ring Puckering.
Chembiochem, 14, 2013
|
|
4LEP
 
 | | Structural insights into substrate recognition in proton dependent oligopeptide transporters | | Descriptor: | N-[(1R)-1-phosphonoethyl]-L-alaninamide, Proton:oligopeptide symporter POT family, ZINC ION | | Authors: | Guettou, F, Quistgaard, E.M, Tresaugues, L, Moberg, P, Jegerschold, C, Zhu, L, Jong, A.J, Nordlund, P, Low, C. | | Deposit date: | 2013-06-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into substrate recognition in proton-dependent oligopeptide transporters.
Embo Rep., 14, 2013
|
|
4HZM
 
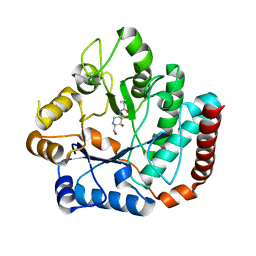 | | Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) bound to N-[(3S,4R,5R,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]butanamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-hexosaminidase, N-[(3S,4R,5R,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]butanamide | | Authors: | Bacik, J.P, Mark, B.L. | | Deposit date: | 2012-11-15 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Development of Selective Inhibitors of NagZ: Increased Susceptibility of Gram-Negative Bacteria to beta-Lactams.
Chembiochem, 14, 2013
|
|
4LHU
 
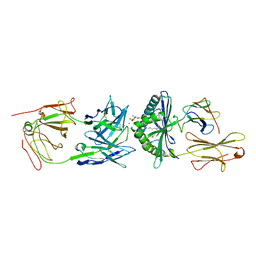 | | Crystal Structure of 9C2 TCR bound to CD1d | | Descriptor: | (15Z)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]tetracos-15-enamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Uldrich, A.P, Le Nours, J, Pellicci, D.G, Gras, S, Rossjohn, J, Godfrey, D.I. | | Deposit date: | 2013-07-01 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | CD1d-lipid antigen recognition by the gamma delta TCR.
Nat.Immunol., 14, 2013
|
|
3EUN
 
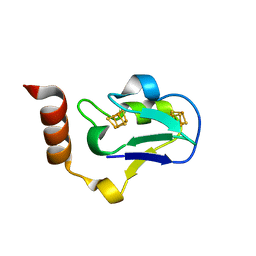 | |
6Z0K
 
 | | Crystal structure of laccase from Pediococcus acidilactici Pp5930 (Hepes pH 7.5) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Putative multicopper oxidase mco | | Authors: | Casino, P, Huesa, J, Pardo, I. | | Deposit date: | 2020-05-09 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis and biochemical properties of laccase enzymes from two Pediococcus species.
Microb Biotechnol, 14, 2021
|
|
6Z0J
 
 | |
1YMP
 
 | |
1Q6K
 
 | | Cathepsin K complexed with t-butyl(1S)-1-cyclohexyl-2-oxoethylcarbamate | | Descriptor: | Cathepsin K, SULFATE ION, TERT-BUTYL(1S)-1-CYCLOHEXYL-2-OXOETHYLCARBAMATE | | Authors: | Catalano, J.G, Deaton, D.N, Furfine, E.S, Hassell, A.M, McFadyen, R.B, Miller, A.B, Miller, L.R, Shewchuk, L.M, Willard, D.H, Wright, L.L. | | Deposit date: | 2003-08-13 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploration of the P1 SAR of aldehyde cathepsin K inhibitors
Bioorg.Med.Chem.Lett., 14, 2004
|
|
2DD9
 
 | | A mutant of GFP-like protein from Chiridius poppei | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CHLORIDE ION, green fluorescent protein | | Authors: | Suto, K, Masuda, H, Takenaka, Y, Mizuno, H. | | Deposit date: | 2006-01-24 | | Release date: | 2007-01-23 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for red-shifted emission of a GFP-like protein from the marine copepod Chiridius poppei
Genes Cells, 14, 2009
|
|
2DD7
 
 | | A GFP-like protein from marine copepod, Chiridius poppei | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CHLORIDE ION, green fluorescent protein | | Authors: | Suto, K, Masuda, H, Takenaka, Y, Mizuno, H. | | Deposit date: | 2006-01-23 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for red-shifted emission of a GFP-like protein from the marine copepod Chiridius poppei
Genes Cells, 14, 2009
|
|
2DU3
 
 | |
1YX7
 
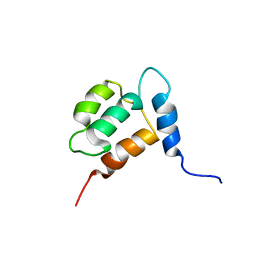 | | NMR structure of Calsensin, energy minimized average structure. | | Descriptor: | Calsensin | | Authors: | Venkitaramani, D.V, Fulton, D.B, Andreotti, A.H, Johansen, K.M, Johansen, J. | | Deposit date: | 2005-02-19 | | Release date: | 2005-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of Calsensin, an invertebrate neuronal calcium-binding protein.
Protein Sci., 14, 2005
|
|
1YZX
 
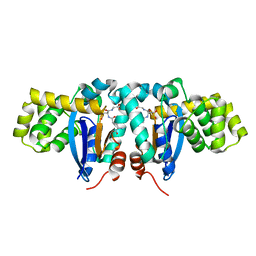 | | Crystal structure of human kappa class glutathione transferase | | Descriptor: | Glutathione S-transferase kappa 1, L-GAMMA-GLUTAMYL-3-SULFINO-L-ALANYLGLYCINE | | Authors: | Li, J, Xia, Z, Ding, J. | | Deposit date: | 2005-02-28 | | Release date: | 2005-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Thioredoxin-like domain of human kappa class glutathione transferase reveals sequence homology and structure similarity to the theta class enzyme
PROTEIN SCI., 14, 2005
|
|
1Z9B
 
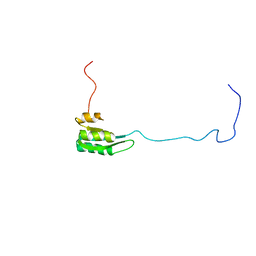 | | Solution structure of the C1-subdomain of Bacillus stearothermophilus translation initiation factor IF2 | | Descriptor: | Translation initiation factor IF-2 | | Authors: | Wienk, H, Tomaselli, S, Bernard, C, Spurio, R, Picone, D, Gualerzi, C.O, Boelens, R. | | Deposit date: | 2005-04-01 | | Release date: | 2005-08-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C1-subdomain of Bacillus stearothermophilus translation initiation factor IF2
Protein Sci., 14, 2005
|
|
2DU7
 
 | |
2DT7
 
 | | Solution structure of the second SURP domain of human splicing factor SF3a120 in complex with a fragment of human splicing factor SF3a60 | | Descriptor: | Splicing factor 3 subunit 1, Splicing factor 3A subunit 3 | | Authors: | He, F, Kuwasako, K, Inoue, M, Guntert, P, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-11 | | Release date: | 2006-12-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the SURP domains and the subunit-assembly mechanism within the splicing factor SF3a complex in 17S U2 snRNP
Structure, 14, 2006
|
|
2DEB
 
 | | Crystal structure of rat carnitine palmitoyltransferase 2 in space group C2221 | | Descriptor: | COENZYME A, Carnitine O-palmitoyltransferase II, mitochondrial, ... | | Authors: | Rufer, A.C, Thoma, R, Benz, J, Stihle, M, Gsell, B, De Roo, E, Banner, D.W, Mueller, F, Chomienne, O, Hennig, M. | | Deposit date: | 2006-02-10 | | Release date: | 2007-02-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of carnitine palmitoyltransferase 2 and implications for diabetes treatment
Structure, 14, 2006
|
|
1ZGZ
 
 | |
2DU4
 
 | |
1ZMV
 
 | |
1ZN5
 
 | |
1ZOW
 
 | | Crystal Structure of S. aureus FabH, beta-ketoacyl carrier protein synthase III | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase III | | Authors: | Qiu, X, Choudhry, A.E, Janson, C.A, Grooms, M, Daines, R.A, Lonsdale, J.T, Khandekar, S.S. | | Deposit date: | 2005-05-15 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and substrate specificity of the beta-ketoacyl-acyl carrier protein synthase III (FabH) from Staphylococcus aureus.
Protein Sci., 14, 2005
|
|
1ZWK
 
 | |
