8DPM
 
 | |
7M07
 
 | | Pre-catalytic ternary complex of DNA Polymerase Lambda with bound 1-nt gapped SSB substrate and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
6ZW6
 
 | | C16 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6M73
 
 | | Crystal structure of Enterococcus hirae L-lactate oxidase in complex with D-lactate form ligand | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Yoshida, H, Hiraka, K, Tsugawa, W, Sode, K. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of lactate oxidase from Enterococcus hirae revealed new aspects of active site loop function: Product-inhibition mechanism and oxygen gatekeeper
Protein Sci., 31, 2022
|
|
6OZ1
 
 | | Crystal structure of the adenylation (A) domain of the carboxylate reductase (CAR) GR01_22995 from Mycobacterium chelonae | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Fedorchuk, T, Khusnutdinova, A, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | One-Pot Biocatalytic Transformation of Adipic Acid to 6-Aminocaproic Acid and 1,6-Hexamethylenediamine Using Carboxylic Acid Reductases and Transaminases.
J.Am.Chem.Soc., 142, 2020
|
|
8DVG
 
 | | Structure of KRAS WT(7-16)-HLA-A*03:01 | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, HLA class I histocompatibility antigen, ... | | Authors: | Wright, K.M, Miller, M, Gabelli, S.B. | | Deposit date: | 2022-07-28 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Hydrophobic interactions dominate the recognition of a KRAS G12V neoantigen.
Nat Commun, 14, 2023
|
|
6UTH
 
 | |
6ZUE
 
 | |
8DME
 
 | | CYP102A1 in Open Conformation | | Descriptor: | 6-methoxy-2-{[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]sulfanyl}-1H-benzimidazole, Bifunctional cytochrome P450/NADPH--P450 reductase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Su, M, Xu, H. | | Deposit date: | 2022-07-08 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Insight into the conformational dynamics of cytochrome P450 CYP102A1 enzyme using Cryo-EM
To Be Published
|
|
6UU6
 
 | | E. coli sigma-S transcription initiation complex with a 4-nt RNA and a UTP ("Old" crystal soaked with UTP, ddCTP, and dinucleotide ApG for 30 minutes) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.201 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
7MMC
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR01-115 | | Descriptor: | (1-methylcyclopropyl)methyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3 protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
6M74
 
 | | Crystal structure of Enterococcus hirae L-lactate oxidase M207L in complex with D-lactate form ligand | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Yoshida, H, Hiraka, K, Tsugawa, W, Sode, K. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure of lactate oxidase from Enterococcus hirae revealed new aspects of active site loop function: Product-inhibition mechanism and oxygen gatekeeper
Protein Sci., 31, 2022
|
|
6OZA
 
 | | Crystal structure of the phycocyanobilin-bound GAF domain from a cyanobacterial phytochrome | | Descriptor: | PHYCOCYANOBILIN, Two-component sensor histidine kinase | | Authors: | Heewhan, S, Xiaoli, Z, Yafang, S, Zhong, R, Wolfgang, G, Kai, H.Z, Xiaojing, Y. | | Deposit date: | 2019-05-15 | | Release date: | 2020-05-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | The interplay between chromophore and protein determines the extended excited state dynamics in a single-domain phytochrome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7MK6
 
 | | KcsA open gate E71V mutant with sodium | | Descriptor: | Fab heavy chain, Fab light chain, pH-gated potassium channel KcsA | | Authors: | Rohaim, A, Li, J, Weingarth, M, Roux, B. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A distinct mechanism of C-type inactivation in the Kv-like KcsA mutant E71V.
Nat Commun, 13, 2022
|
|
8DNR
 
 | |
6ZZX
 
 | | Structure of low-light grown Chlorella ohadii Photosystem I | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (3R)-beta,beta-caroten-3-ol, ... | | Authors: | Caspy, I, Nelson, N, Nechushtai, R, Neumann, E, Shkolnisky, Y. | | Deposit date: | 2020-08-05 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
6PI8
 
 | |
6V0H
 
 | | Lipophilic Envelope-spanning Tunnel B (LetB), Model 6 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Isom, G.L, Coudray, N, MacRae, M.R, McManus, C.T, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2019-11-18 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope.
Cell, 181, 2020
|
|
6M6X
 
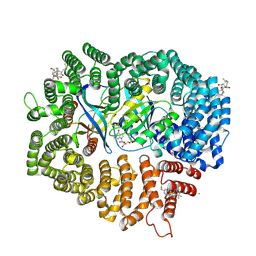 | | Oridonin in complex with CRM1#-Ran-RanBP1 | | Descriptor: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Novel Mechanistic Observations and NES-Binding Groove Features Revealed by the CRM1 Inhibitors Plumbagin and Oridonin.
J.Nat.Prod., 84, 2021
|
|
8E8Z
 
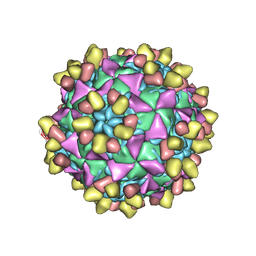 | | 9H2 Fab-Sabin poliovirus 1 complex | | Descriptor: | 9H2 Fab heavy chain, 9H2 Fab light chain, Capsid protein VP1, ... | | Authors: | Charnesky, A.J. | | Deposit date: | 2022-08-26 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | A human monoclonal antibody binds within the poliovirus receptor-binding site to neutralize all three serotypes.
Nat Commun, 14, 2023
|
|
7MHR
 
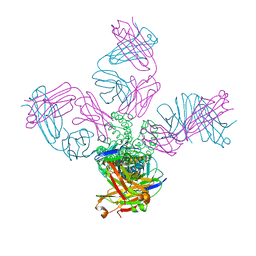 | | KcsA E71V closed gate with K+ | | Descriptor: | Fab heavy chain, Fab light chain, POTASSIUM ION, ... | | Authors: | Rohaim, A, Li, J, Weingarth, M, Roux, B. | | Deposit date: | 2021-04-15 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | A distinct mechanism of C-type inactivation in the Kv-like KcsA mutant E71V.
Nat Commun, 13, 2022
|
|
6V11
 
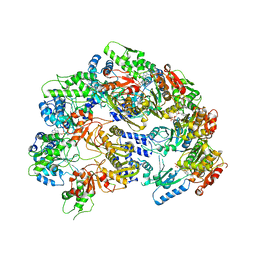 | | Lon Protease from Yersinia pestis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Shin, M, Puchades, C, Asmita, A, Puri, N, Adjei, E, Wiseman, R.L, Karzai, A.W, Lander, G.C. | | Deposit date: | 2019-11-19 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for distinct operational modes and protease activation in AAA+ protease Lon.
Sci Adv, 6, 2020
|
|
6ZW7
 
 | | C17 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6PIK
 
 | | Tetrameric cryo-EM ArnA | | Descriptor: | Bifunctional polymyxin resistance protein ArnA, UDP-4-amino-4-deoxy-L-arabinose formyltransferase | | Authors: | Yang, M, Gehring, K. | | Deposit date: | 2019-06-26 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Cryo-electron microscopy structures of ArnA, a key enzyme for polymyxin resistance, revealed unexpected oligomerizations and domain movements.
J.Struct.Biol., 208, 2019
|
|
6LU5
 
 | | Crystal structure of BPTF-BRD with ligand DCBPin5 bound | | Descriptor: | 6-(1H-indol-5-yl)-N-methyl-2-methylsulfonyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2020-01-25 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86527729 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
