6TOJ
 
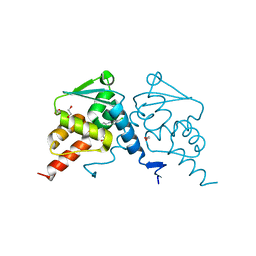 | | Crystal structure of human BCL6 BTB domain in complex with compound 17a | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[1-methyl-3-(3-methyl-3-oxidanyl-butyl)-2-oxidanylidene-benzimidazol-5-yl]amino]pyridine-3-carbonitrile, B-cell lymphoma 6 protein | | Authors: | Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2019-12-11 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | AchievingIn VivoTarget Depletion through the Discovery and Optimization of Benzimidazolone BCL6 Degraders.
J.Med.Chem., 63, 2020
|
|
7BBX
 
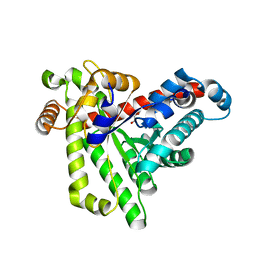 | | Neisseria gonorrhoeae transaldolase, variant K8A | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Transaldolase | | Authors: | Rabe von Pappenheim, F, Wensien, M, Funk, L.M, Tittmann, K. | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | A lysine-cysteine redox switch with an NOS bridge regulates enzyme function.
Nature, 593, 2021
|
|
9FZI
 
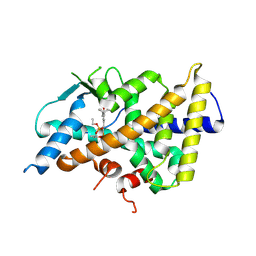 | | A new crystal structure of the hPXR-LBD in fusion with an SRC1 co-activator peptide and in complex with SR12813 (P212121 form) | | Descriptor: | Nuclear receptor subfamily 1 group I member 2,Nuclear receptor coactivator 1, [2-(3,5-DI-TERT-BUTYL-4-HYDROXY-PHENYL)-1-(DIETHOXY-PHOSPHORYL)-VINYL]-PHOSPHONIC ACID DIETHLYL ESTER | | Authors: | Carivenc, C, Blanc, P, Bourguet, W, Delfosse, V. | | Deposit date: | 2024-07-05 | | Release date: | 2025-02-19 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A two-in-one expression construct for biophysical and structural studies of the human pregnane X receptor ligand-binding domain, a pharmaceutical and environmental target.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
8SSO
 
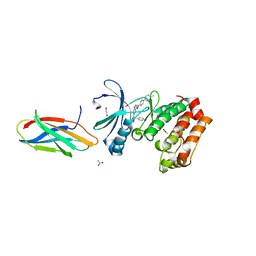 | | AurA bound to danusertib and inhibiting monobody Mb2 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Aurora kinase A, ... | | Authors: | Ludewig, H, Kim, C, Kern, D. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A biophysical framework for double-drugging kinases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
9BH5
 
 | | High-resolution C. elegans 80S ribosome structure - class 1 | | Descriptor: | 18S rRNA, 28S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Sehgal, E, Serrao, V.H.B, Arribere, J. | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | High-resolution reconstruction of a C. elegans ribosome sheds light on evolutionary dynamics and tissue specificity.
Rna, 30, 2024
|
|
6G6R
 
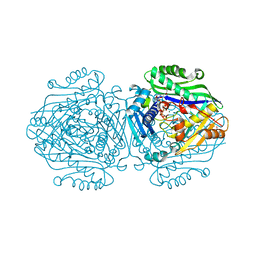 | | Human Methionine Adenosyltransferase II with SAMe and PPNP | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Panmanee, J, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2018-04-02 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Control and regulation of S-Adenosylmethionine biosynthesis by the regulatory beta subunit and quinolone-based compounds.
Febs J., 286, 2019
|
|
5I2F
 
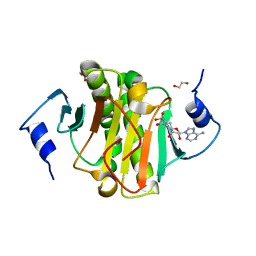 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) with bound sulfamide inhibitor Bio-AMS | | Descriptor: | 1,2-ETHANEDIOL, 5'-deoxy-5'-[({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}sulfamoyl)amino]adenosine, Histidine triad nucleotide-binding protein 1 | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Design, Synthesis, and Characterization of Sulfamide and Sulfamate Nucleotidomimetic Inhibitors of hHint1.
Acs Med.Chem.Lett., 7, 2016
|
|
5NXV
 
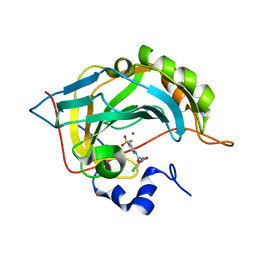 | | Carbonic Anhydrase II Inhibitor RA8 | | Descriptor: | 2-(4-chloranyl-5-methyl-3-nitro-pyrazol-1-yl)-~{N}-(4-sulfamoylphenyl)ethanamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
3O84
 
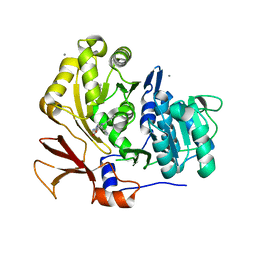 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 6-phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 6-phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid, ... | | Authors: | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
5O33
 
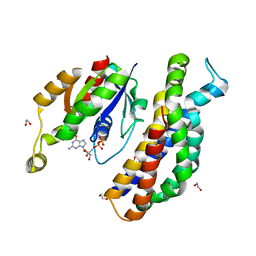 | | A structure of the GEF Kalirin DH1 domain in complex with the small GTPase Rac1 | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, Kalirin, ... | | Authors: | Gray, J, Krojer, T, Talon, R, Fairhead, M, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P, von Delft, F. | | Deposit date: | 2017-05-23 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A structure of the GEF Kalirin DH1 domain in complex with the small GTPase Rac1
To Be Published
|
|
3JVR
 
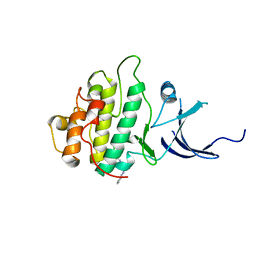 | | Characterization of the Chk1 allosteric inhibitor binding site | | Descriptor: | (1S)-1-(1H-benzimidazol-2-yl)ethyl (3,4-dichlorophenyl)carbamate, Serine/threonine-protein kinase Chk1 | | Authors: | Chen, P. | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Characterization of the CHK1 allosteric inhibitor binding site.
Biochemistry, 48, 2009
|
|
7YZZ
 
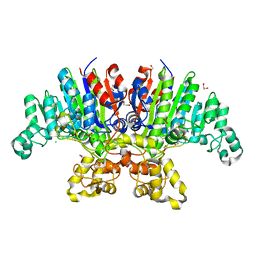 | | Crystal structure of Vibrio alkaline phosphatase in 0.5 M NaCl | | Descriptor: | 1,2-ETHANEDIOL, Alkaline phosphatase, CHLORIDE ION, ... | | Authors: | Markusson, S, Hjorleifsson, J.G, Kursula, P, Asgeirsson, B. | | Deposit date: | 2022-02-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structural Characterization of Functionally Important Chloride Binding Sites in the Marine Vibrio Alkaline Phosphatase.
Biochemistry, 61, 2022
|
|
7FY9
 
 | | Crystal Structure of human FABP4 binding site mutated to that of FABP5 in complex with 2-cyclopentyl-4-(4-fluorophenyl)-6-[1-(methoxymethyl)cyclopentyl]-3-methyl-5-(1H-tetrazol-5-yl)pyridine, i.e. SMILES c1(c(nc(c(c1c1ccc(cc1)F)C1=NN=NN1)C1(CCCC1)COC)C1CCCC1)C with IC50=0.14164 microM | | Descriptor: | (5P)-2-cyclopentyl-4-(4-fluorophenyl)-6-[1-(methoxymethyl)cyclopentyl]-3-methyl-5-(1H-tetrazol-5-yl)pyridine, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Kuhne, H, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
6BSX
 
 | |
8Y01
 
 | | Cryo-EM structure of Medium-wave-sensitive opsin 1 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Peng, Q, Jiang, H.H, Cheng, X.Y, Li, J, Zhang, J. | | Deposit date: | 2024-01-21 | | Release date: | 2025-02-12 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Cryo-EM structure of Medium-wave-sensitive opsin 1
To Be Published
|
|
7FWC
 
 | | Crystal Structure of human FABP4 in complex with 5-(4-chlorophenyl)-2-(hydroxymethylene)cyclohexane-1,3-dione | | Descriptor: | 5-(4-chlorophenyl)-2-(hydroxymethylene)cyclohexane-1,3-dione, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
6RU8
 
 | | Crystal structure of Casein Kinase I delta (CK1d) in complex with triple phosphorylated p63 PAD3P peptide | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Casein kinase I isoform delta, ... | | Authors: | Chaikuad, A, Tuppi, M, Gebel, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Dotsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | p63 uses a switch-like mechanism to set the threshold for induction of apoptosis.
Nat.Chem.Biol., 16, 2020
|
|
7BHU
 
 | | Crystal structure of MAT2a with elaborated fragment 26 bound in the allosteric site | | Descriptor: | 1,2-ETHANEDIOL, 7-chloranyl-4-(dimethylamino)-1-(2-hydroxyethyl)quinazolin-2-one, S-ADENOSYLMETHIONINE, ... | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
8EWL
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, {N-[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]-3-(pyridin-4-yl)propanamide}bis[2-(quinolin-2-yl-kappaN)phenyl-kappaC~1~]iridium(1+) | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-10-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Dynamic Ir(III) Photosensors for the Major Human Drug-Metabolizing Enzyme Cytochrome P450 3A4.
Inorg.Chem., 62, 2023
|
|
7FZR
 
 | | Crystal Structure of human FABP4 in complex with 4-hydroxy-6-(2-naphthalen-1-ylethyl)pyran-2-one, i.e. SMILES c1ccc2c(c1)c(ccc2)CCC1=CC(=CC(=O)O1)O with IC50=0.183 microM | | Descriptor: | 4-hydroxy-6-[2-(naphthalen-1-yl)ethyl]-2H-pyran-2-one, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Maurer, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
9IX7
 
 | | Crystal structure of homolog of dihydroxyacid dehydratase(AstD) from Aspergillus terreus | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, dihydroxy-acid dehydratase | | Authors: | Huang, W.X, Zhang, P.X, Zhou, J.H. | | Deposit date: | 2024-07-26 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Bases of Dihydroxy Acid Dehydratase Inhibition and Biodesign for Self-Resistance.
Biodes Res, 6, 2024
|
|
6BXR
 
 | | Crystal structure of Toxoplasma gondii Mitochondrial Association Factor 1 B (MAF1B) | | Descriptor: | BROMIDE ION, GLYCEROL, Mitochondrial association factor 1, ... | | Authors: | Parker, M.L, Ramaswamy, R, Boulanger, M.J. | | Deposit date: | 2017-12-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Toxoplasma gondii locus required for the direct manipulation of host mitochondria has maintained multiple ancestral functions.
Mol. Microbiol., 108, 2018
|
|
6RSD
 
 | |
7FZT
 
 | | Crystal Structure of human FABP4 binding site mutated to that of FABP5 in complex with rac-(1R,2S)-2-[(3,4-dichlorobenzoyl)amino]cyclohexane-1-carboxylic acid, i.e. SMILES C1CC[C@@H]([C@@H](C1)C(=O)O)NC(=O)c1cc(c(cc1)Cl)Cl with IC50=15.8182 microM | | Descriptor: | (1R,2S)-2-(3,4-dichlorobenzamido)cyclohexane-1-carboxylic acid, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Buettelmann, B, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7G1Y
 
 | | Crystal Structure of human FABP4 in complex with 2-[(3-methoxyphenyl)sulfanylmethyl]-1,3-thiazole-4-carboxylic acid | | Descriptor: | 2-{[(3-methoxyphenyl)sulfanyl]methyl}-1,3-thiazole-4-carboxylic acid, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Grenz-Achim, K, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
