7MSQ
 
 | |
1ENE
 
 | |
7MOH
 
 | | Crystal Structure of Arabidopsis thaliana Plant and Fungi Atypical Dual Specificity Phosphatase 1(AtPFA-DSP1 ) Cys150Ser in complex with 5-diphosphoinositol 1,3,4,6-tetrakisphosphate (5PP-InsP4) and phosphate in conformation B (Pi(B)) | | Descriptor: | (1r,2R,3S,4r,5R,6S)-4-hydroxy-2,3,5,6-tetrakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, PHOSPHATE ION, Tyrosine-protein phosphatase DSP1 | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2021-05-01 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural expose of noncanonical molecular reactivity within the protein tyrosine phosphatase WPD loop.
Nat Commun, 13, 2022
|
|
5WFJ
 
 | | THE JAK3 KINASE DOMAIN IN COMPLEX WITH A COVALENT INHIBITOR | | Descriptor: | 4-({[3-(propanoylamino)phenyl]methyl}amino)pyrrolo[1,2-b]pyridazine-3-carboxamide, Tyrosine-protein kinase JAK3 | | Authors: | Sack, J. | | Deposit date: | 2017-07-12 | | Release date: | 2017-10-04 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of highly potent, selective, covalent inhibitors of JAK3.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
1EN3
 
 | |
1EN8
 
 | |
4K4U
 
 | | Poliovirus polymerase elongation complex (r5_form) | | Descriptor: | RNA (5'-R(*AP*AP*GP*UP*CP*UP*CP*CP*AP*GP*GP*UP*CP*UP*CP*UP*CP*UP*CP*GP*UP*CP*GP*AP*AP*A)-3'), RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*GP*A)-3'), RNA (5'-R(P*GP*GP*GP*GP*GP*AP*GP*AP*UP*GP*A)-3'), ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2013-04-12 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts.
Plos One, 8, 2013
|
|
7MOL
 
 | |
4AUN
 
 | | Crystal structure, recombinant expression and mutagenesis studies of the bifunctional catalase-phenol oxidase from Scytalidium thermophilum | | Descriptor: | CALCIUM ION, CATALASE-PHENOL OXIDASE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE | | Authors: | Yuzugullu, Y, Trinh, C.H, Smith, M.A, Pearson, A.R, Phillips, S.E.V, Sutay Kocabas, D, Bakir, U, Ogel, Z.B, McPherson, M.J. | | Deposit date: | 2012-05-18 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure, Recombinant Expression and Mutagenesis Studies of the Catalase with Oxidase Activity from Scytalidium Thermophilum
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BCS
 
 | | Crystal structure of an avidin mutant | | Descriptor: | ACETATE ION, BIOTIN, CHIMERIC AVIDIN, ... | | Authors: | Airenne, T.T, Niederhauser, B, Hytonen, V.P, Kulomaa, M.S, Johnson, M.S. | | Deposit date: | 2012-10-03 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Novel Chimeric Avidin with Increased Thermal Stability Using DNA Shuffling.
Plos One, 9, 2014
|
|
8BLP
 
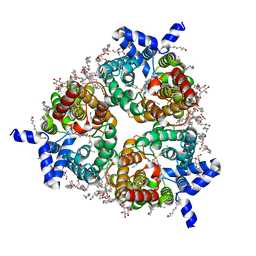 | | Human Urea Transporter UT-B/UT1 in Complex with Inhibitor UTBinh-14 | | Descriptor: | 10-(4-ethylphenyl)sulfonyl-~{N}-(thiophen-2-ylmethyl)-5-thia-1,8,11,12-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-2(6),3,7,9,11-pentaen-7-amine, CHOLESTEROL HEMISUCCINATE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Chi, G, Dietz, L, Pike, A.C.W, Maclean, E.M, Mukhopadhyay, S.M.M, Bohstedt, T, Wang, D, Scacioc, A, McKinley, G, Arrowsmith, C.H, Edwards, A, Bountra, C, Fernandez-Cid, A, Burgess-Brown, N.A, Duerr, K.L. | | Deposit date: | 2022-11-10 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural characterization of human urea transporters UT-A and UT-B and their inhibition.
Sci Adv, 9, 2023
|
|
3M8Y
 
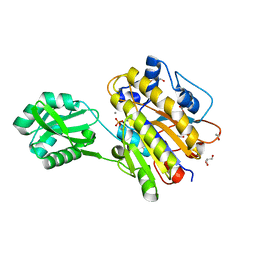 | | Phosphopentomutase from Bacillus cereus after glucose-1,6-bisphosphate activation | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Panosian, T.D, Nannemann, D.P, Watkins, G, Wadzinski, B, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2010-03-19 | | Release date: | 2010-12-29 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacillus cereus Phosphopentomutase Is an Alkaline Phosphatase Family Member That Exhibits an Altered Entry Point into the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
4K4X
 
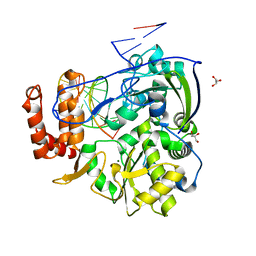 | | Coxsackievirus B3 polymerase elongation complex (r2_form), rna | | Descriptor: | GLYCEROL, MAGNESIUM ION, RNA (5'-R(*AP*AP*GP*UP*CP*UP*CP*CP*AP*GP*GP*UP*CP*UP*CP*UP*CP*GP*UP*CP*GP*AP*AP*A)-3'), ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2013-04-12 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts.
Plos One, 8, 2013
|
|
2MND
 
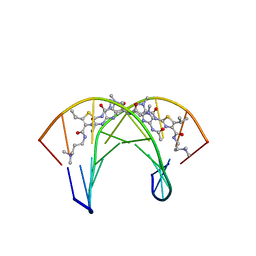 | | Recognition complex of DNA d(CGACGCGTCG)2 with thiazotropsin B | | Descriptor: | 5'-D(*CP*GP*AP*CP*GP*CP*GP*TP*CP*G)-3', thiazotropsin B | | Authors: | Alniss, H.Y, Salvia, M.-V, Sadikov, M, Golovchenko, I, Anthony, N.G, Khalaf, A.I, Mackay, S.P, Suckling, C.J, Parkinson, J.A. | | Deposit date: | 2014-04-03 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the DNA minor groove by thiazotropsin analogues.
Chembiochem, 15, 2014
|
|
6WPT
 
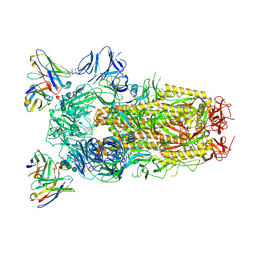 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the S309 neutralizing antibody Fab fragment (open state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, ... | | Authors: | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, De Marco, A, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-27 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
7M6K
 
 | |
2LWO
 
 | | Solution Structure of Duplex DNA Containing a b-Carba-Fapy-dG Lesion | | Descriptor: | DNA (5'-D(*G*CP*GP*TP*AP*C*(LWM)P*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*GP*TP*AP*CP*G)-3') | | Authors: | Zalianyak, T, de los Santos, C, Lukin, M, Attaluri, S, Johnson, F. | | Deposit date: | 2012-08-03 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Duplex DNA Containing a b-Carba-Fapy-dG Lesion
Chem.Res.Toxicol., 25, 2012
|
|
2LWN
 
 | | Solution Structure of Duplex DNA Containing a b-Carba-Fapy-dG Lesion | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*C*(LWM)P*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*GP*TP*AP*CP*G)-3') | | Authors: | Zalianyak, T, de los Santos, C, Lukin, M, Attaluri, S, Johnson, F. | | Deposit date: | 2012-08-03 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Duplex DNA Containing a b-Carba-Fapy-dG Lesion
Chem.Res.Toxicol., 25, 2012
|
|
2LWM
 
 | | Solution Structure of Duplex DNA Containing a b-Carba-Fapy-dG Lesion | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*C*(LWM)P*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*GP*TP*AP*CP*G)-3') | | Authors: | Zalianyak, T, de los Santos, C, Lukin, M, Attaluri, S, Johnson, F. | | Deposit date: | 2012-08-03 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Duplex DNA Containing a b-Carba-Fapy-dG Lesion
Chem.Res.Toxicol., 25, 2012
|
|
4K4V
 
 | | Poliovirus polymerase elongation complex (r5+1_form) | | Descriptor: | DNA/RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*GP*A)-D(P*C)-3'), RNA (5'-R(*AP*AP*GP*UP*CP*UP*CP*CP*AP*GP*GP*UP*CP*UP*CP*UP*CP*UP*CP*GP*UP*CP*GP*AP*AP*A)-3'), RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2013-04-12 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts.
Plos One, 8, 2013
|
|
8BHM
 
 | | GABA-A receptor a5 homomer - a5V3 - DMCM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5, methyl 4-ethyl-6,7-dimethoxy-9H-pyrido[3,4-b]indole-3-carboxylate | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6QX2
 
 | | 3.4A structure of benzoisoxazole 3 with S.aureus DNA gyrase and DNA | | Descriptor: | (2~{R})-2-[[5-(2-chlorophenyl)-1,2-benzoxazol-3-yl]oxy]-2-phenyl-ethanamine, DNA (5'-D(*GP*AP*GP*CP*GP*TP*AP*CP*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D. | | Deposit date: | 2019-03-06 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure-guided design of antibacterials that allosterically inhibit DNA gyrase.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
4KIK
 
 | | Human IkB kinase beta | | Descriptor: | Inhibitor of nuclear factor kappa-B kinase subunit beta, K-252A | | Authors: | Liu, S, Mosyak, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-06-26 | | Last modified: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Crystal Structure of a Human I kappa B Kinase beta Asymmetric Dimer.
J.Biol.Chem., 288, 2013
|
|
1ELA
 
 | | Analogous inhibitors of elastase do not always bind analogously | | Descriptor: | 6-ammonio-N-(trifluoroacetyl)-L-norleucyl-N-[4-(1-methylethyl)phenyl]-L-prolinamide, ACETIC ACID, CALCIUM ION, ... | | Authors: | Mattos, C, Rasmussen, B, Ding, X, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-04-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analogous inhibitors of elastase do not always bind analogously.
Nat.Struct.Biol., 1, 1994
|
|
6WPS
 
 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, ... | | Authors: | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, De Marco, A, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-27 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
