4V4R
 
 | | Crystal structure of the whole ribosomal complex. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2005-09-30 | | Release date: | 2014-07-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (5.9 Å) | | Cite: | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
4V8J
 
 | | Crystal structure of the bacterial ribosome ram mutation G347U. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fagan, C.E, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2011-12-20 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Reorganization of an intersubunit bridge induced by disparate 16S ribosomal ambiguity mutations mimics an EF-Tu-bound state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4V4S
 
 | | Crystal structure of the whole ribosomal complex. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2005-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (6.76 Å) | | Cite: | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
5CXF
 
 | |
1LMP
 
 | |
5JWZ
 
 | |
4W6W
 
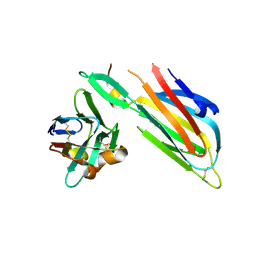 | | Co-complex structure of the lectin domain of F18 fimbrial adhesin FedF with inhibitory nanobody NbFedF6 | | Descriptor: | F18 fimbrial adhesin AC, NbFedF6 | | Authors: | Moonens, K, De Kerpel, M, Annelies, C, Cox, E, Pardon, E, Remaut, H, De Greve, H. | | Deposit date: | 2014-08-21 | | Release date: | 2014-12-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Nanobody Mediated Inhibition of Attachment of F18 Fimbriae Expressing Escherichia coli.
Plos One, 9, 2014
|
|
4W6X
 
 | | Co-complex structure of the lectin domain of F18 fimbrial adhesin FedF with inhibitory nanobody NbFedF7 | | Descriptor: | F18 fimbrial adhesin AC, Nanobody NbFedF7 | | Authors: | Moonens, K, De Kerpel, M, Coddens, A, Cox, E, Pardon, E, Remaut, H, De Greve, H. | | Deposit date: | 2014-08-21 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Nanobody Mediated Inhibition of Attachment of F18 Fimbriae Expressing Escherichia coli.
Plos One, 9, 2014
|
|
7S53
 
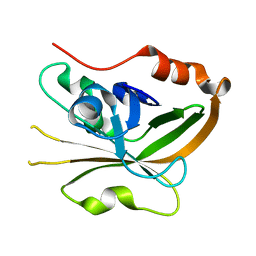 | | Structure of Sortase A from Streptococcus pyogenes with the b7-b8 loop sequence from Listeria monocytogenes Sortase A | | Descriptor: | Class A sortase, sortase A chimera | | Authors: | Johnson, D.A, Svendsen, J.E, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-09-09 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and biochemical analyses of selectivity determinants in chimeric Streptococcus Class A sortase enzymes.
Protein Sci., 31, 2022
|
|
7RQD
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site deacylated tRNA analog CACCA, P-site MTI-tripeptidyl-tRNA analog ACCA-ITM, and chloramphenicol at 2.50A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1UBS
 
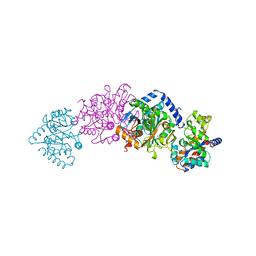 | | TRYPTOPHAN SYNTHASE (E.C.4.2.1.20) WITH A MUTATION OF LYS 87->THR IN THE B SUBUNIT AND IN THE PRESENCE OF LIGAND L-SERINE | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE, SODIUM ION, ... | | Authors: | Rhee, S, Parris, K, Ahmed, S.A, Miles, E.W, Davies, D.R. | | Deposit date: | 1995-12-14 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes.
Biochemistry, 36, 1997
|
|
5JJ5
 
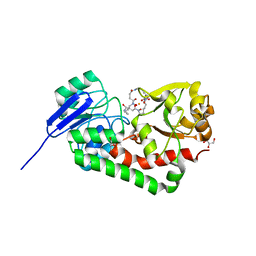 | | Crystal structure of iron uptake ABC transporter substrate-binding protein PiaA from Streptococcus pneumoniae Canada MDR_19A bound to hydroxymate siderophore ferrioxamine E and iron(III) | | Descriptor: | (8E)-6,17,28-trihydroxy-1,6,12,17,23,28-hexaazacyclotritriacont-8-ene-2,5,13,16,24,27-hexone, ABC transporter substrate-binding protein-iron transport, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Kurdritska, M, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-04-22 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of iron uptake ABC transporter substrate-binding protein PiaA from Streptococcus pneumoniae Canada MDR_19A bound to hydroxymate siderophore ferrioxamine E and iron(III)
To Be Published
|
|
1KJZ
 
 | |
1KQJ
 
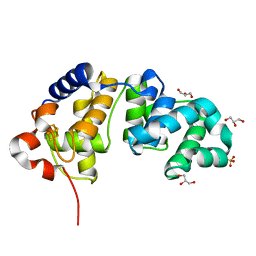 | | Crystal Structure of a Mutant of MutY Catalytic Domain | | Descriptor: | A/G-SPECIFIC ADENINE GLYCOSYLASE, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Messick, T.E, Chmiel, N.H, Golinelli, M.P, David, S.S, Joshua-Tor, L. | | Deposit date: | 2002-01-06 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Noncysteinyl coordination to the [4Fe-4S]2+ cluster of the DNA repair adenine glycosylase MutY introduced via site-directed mutagenesis. Structural characterization of an unusual histidinyl-coordinated cluster.
Biochemistry, 41, 2002
|
|
2HXR
 
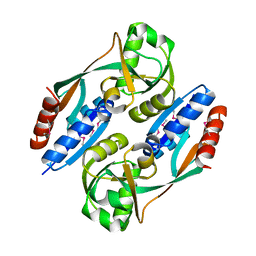 | | Structure of the ligand binding domain of E. coli CynR, a transcriptional regulator controlling cyanate metabolism | | Descriptor: | HTH-type transcriptional regulator cynR | | Authors: | Singer, A.U, Cuff, M.E, Evdokimova, E, Kagan, O, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-03 | | Release date: | 2006-08-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of CynR, a transcriptional regulator, both in the presence and absence of sodium azide, its activator ligand
To be Published
|
|
3SGT
 
 | |
1KMS
 
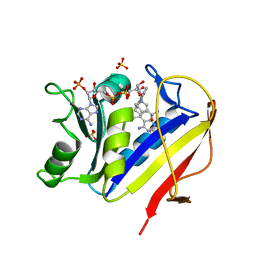 | | HUMAN DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADPH AND 6-([5-QUINOLYLAMINO]METHYL)-2,4-DIAMINO-5-METHYLPYRIDO[2,3-D]PYRIMIDINE (SRI-9439), A LIPOPHILIC ANTIFOLATE | | Descriptor: | 6-([5-QUINOLYLAMINO]METHYL)-2,4-DIAMINO-5-METHYLPYRIDO[2,3-D]PYRIMIDINE, DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Klon, A.E, Heroux, A, Ross, L.J, Pathak, V, Johnson, C.A, Piper, J.R, Borhani, D.W. | | Deposit date: | 2001-12-17 | | Release date: | 2002-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Atomic structures of human dihydrofolate reductase complexed with NADPH and two lipophilic antifolates at 1.09 a and 1.05 a resolution.
J.Mol.Biol., 320, 2002
|
|
4CKL
 
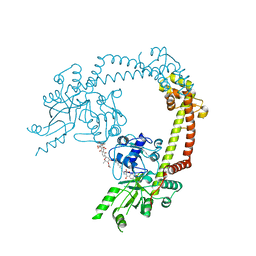 | | Structure of 55 kDa N-terminal domain of E. coli DNA gyrase A subunit with simocyclinone D8 bound | | Descriptor: | DNA GYRASE SUBUNIT A, SIMOCYCLINONE D8 | | Authors: | Hearnshaw, S.J, Edwards, M.J, Stevenson, C.E.M, Lawson, D.M, Maxwell, A. | | Deposit date: | 2014-01-07 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A New Crystal Structure of the Bifunctional Antibiotic Simocyclinone D8 Bound to DNA Gyrase Gives Fresh Insight Into the Mechanism of Inhibition.
J.Mol.Biol., 426, 2014
|
|
4CHS
 
 | | Crystal structure of a tau class glutathione transferase 10 from Glycine max | | Descriptor: | ACETONE, GLUTATHIONE S-TRANSFERASE, S-Hydroxy-Glutathione | | Authors: | Skopelitou, K, Muleta, A.W, Papageorgiou, A.C, Pavli, O, Flemetakis, E, Chronopoulou, E, Skaracis, G.N, Labrou, N.E. | | Deposit date: | 2013-12-04 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Catalytic features and crystal structure of a tau class glutathione transferase from Glycine max specifically upregulated in response to soybean mosaic virus infections.
Biochim. Biophys. Acta, 1854, 2015
|
|
2VER
 
 | | Structural model for the complex between the Dr adhesins and carcinoembryonic antigen (CEA) | | Descriptor: | AFIMBRIAL ADHESIN AFA-III, ARCINOEMBRYONIC ANTIGEN-RELATED CELL ADHESION MOLECULE 5, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Korotkova, N, Yang, Y, Le Trong, I, Cota, E, Demeler, B, Marchant, J, Thomas, W.E, Stenkamp, R.E, Moseley, S.L, Matthews, S. | | Deposit date: | 2007-10-26 | | Release date: | 2008-01-08 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Binding of Dr Adhesins of Escherichia Coli to Carcinoembryonic Antigen Triggers Receptor Dissociation.
Mol.Microbiol., 67, 2008
|
|
8RQU
 
 | | Structure of TEM1 beta-lactamase variant 70.a | | Descriptor: | Beta-lactamase TEM-1, MAGNESIUM ION | | Authors: | Napier, E, Fram, B.F, Gauthier, N.P, Sander, C, Khan, A.R. | | Deposit date: | 2024-01-19 | | Release date: | 2024-02-14 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Simultaneous enhancement of multiple functional properties using evolution-informed protein design.
Nat Commun, 15, 2024
|
|
2V7Q
 
 | | The structure of F1-ATPase inhibited by I1-60HIS, a monomeric form of the inhibitor protein, IF1. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP SYNTHASE DELTA CHAIN, ... | | Authors: | Gledhill, J.R, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2007-07-31 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | How the Regulatory Protein, If1, Inhibits F1- ATPase from Bovine Mitochondria.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2I5Q
 
 | | Crystal structure of Apo L-rhamnonate dehydratase from Escherichia Coli | | Descriptor: | L-rhamnonate dehydratase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-25 | | Release date: | 2006-09-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: L-rhamnonate dehydratase.
Biochemistry, 47, 2008
|
|
3SGV
 
 | |
3SGX
 
 | |
