4WEL
 
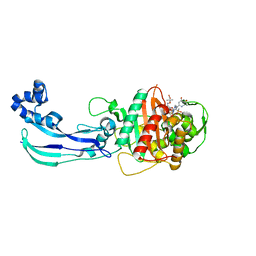 | | Crystal structure of Pseudomonas aeruginosa PBP3 with SMC-3176 | | Descriptor: | (3S,4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-4-formyl-1-[({3-(5-hydroxy-4-oxo-3,4-dihydropyridin-2-yl)-4-[3-(methylsulfonyl)propyl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-3,10,10-trimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oic acid, Penicillin-binding protein 3 | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | SAR and Structural Analysis of Siderophore-Conjugated Monocarbam Inhibitors of Pseudomonas aeruginosa PBP3.
Acs Med.Chem.Lett., 6, 2015
|
|
4WEK
 
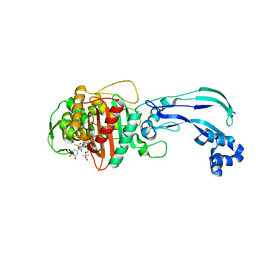 | | Crystal structure of Pseudomonas aeruginosa PBP3 with a R4 substituted vinyl monocarbam | | Descriptor: | (3S,4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-3-ethenyl-4-formyl-1-[({3-(5-hydroxy-4-oxo-3,4-dihydropyridin-2-yl)-4-[3-(methylsulfonyl)propyl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-10,10-dimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oic acid, Penicillin-binding protein 3 | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | SAR and Structural Analysis of Siderophore-Conjugated Monocarbam Inhibitors of Pseudomonas aeruginosa PBP3.
Acs Med.Chem.Lett., 6, 2015
|
|
8P1U
 
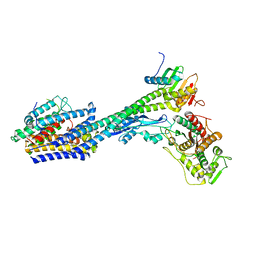 | | Structure of divisome complex FtsWIQLB | | Descriptor: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ, ... | | Authors: | Yang, L, Chang, S, Tang, D, Dong, H. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the activation of the divisome complex FtsWIQLB.
Cell Discov, 10, 2024
|
|
6UN3
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with ticarcillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-carboxy-2-(thiophen-3-yl)acetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CALCIUM ION, GLYCEROL, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2019-10-10 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Influence of the alpha-Methoxy Group on the Reaction of Temocillin with Pseudomonas aeruginosa PBP3 and CTX-M-14 beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2019
|
|
6UN1
 
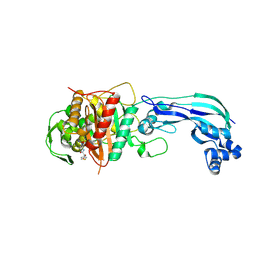 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with temocillin | | Descriptor: | (2R,4S)-2-[(1S)-1-{[(2R)-2-carboxy-2-(thiophen-3-yl)acetyl]amino}-1-methoxy-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4 -carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2019-10-10 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Influence of the alpha-Methoxy Group on the Reaction of Temocillin with Pseudomonas aeruginosa PBP3 and CTX-M-14 beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2019
|
|
6G0K
 
 | | Crystal structure of Enterococcus faecium D63r Penicillin-Binding protein 5 (PBP5fm) | | Descriptor: | Low affinity penicillin-binding protein 5 (PBP5), SULFATE ION | | Authors: | Sauvage, E, El Gachi, M, Herman, R, Kerff, F, Charlier, P. | | Deposit date: | 2018-03-19 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of inactivation of Enterococcus faecium penicillin binding protein 5 by ceftobiprole.
To Be Published
|
|
7ZG8
 
 | |
6G9P
 
 | | Structural basis for the inhibition of E. coli PBP2 | | Descriptor: | Peptidoglycan D,D-transpeptidase MrdA | | Authors: | Ruff, M, Levy, N. | | Deposit date: | 2018-04-11 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural Basis for E. coli Penicillin Binding Protein (PBP) 2 Inhibition, a Platform for Drug Design.
J.Med.Chem., 62, 2019
|
|
7ZUK
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) in complex with lactone 7Az - Streptococcus pneumoniae R6 | | Descriptor: | 6-azido-N-[(2S)-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]amino]-3-phenyl-propan-2-yl]hexanamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
7ZUJ
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) in complex with lactone 6Az - Streptococcus pneumoniae R6 | | Descriptor: | 6-azido-N-[(2S)-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]amino]propan-2-yl]hexanamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
7ZUL
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) in complex with 8Az lactone - Streptococcus pneumoniae R6 | | Descriptor: | 6-azido-N-[(2R)-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]amino]-3-phenyl-propan-2-yl]hexanamide, CHLORIDE ION, Penicillin-binding protein 1b | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.744 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
6G9F
 
 | | Structural basis for the inhibition of E. coli PBP2 | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Peptidoglycan D,D-transpeptidase MrdA | | Authors: | Ruff, M, Levy, N. | | Deposit date: | 2018-04-10 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for E. coli Penicillin Binding Protein (PBP) 2 Inhibition, a Platform for Drug Design.
J.Med.Chem., 62, 2019
|
|
7ZUI
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) in complex with lactone 5Az - Streptococcus pneumoniae R6 | | Descriptor: | 6-azido-N-[(2R)-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]amino]propan-2-yl]hexanamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
7ZUH
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) Streptococcus pneumoniae R6 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Penicillin-binding protein 1b | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.467 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
6G88
 
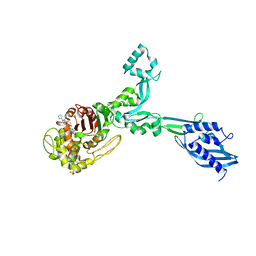 | | Crystal structure of Enterococcus Faecium D63r Penicillin-Binding protein 5 (PBP5fm) | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyrrolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Low affinity penicillin-binding protein 5 (PBP5), SULFATE ION | | Authors: | Sauvage, E, El Gachi, M, Herman, R, Kerff, F, Charlier, P. | | Deposit date: | 2018-04-08 | | Release date: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of inactivation of Enterococcus faecium penicillin binding protein 5 by ceftobiprole.
To Be Published
|
|
6BSR
 
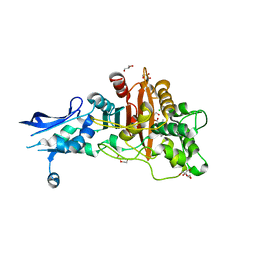 | | Crystal structure of penicillin-binding protein 4 (PBP4) from Enterococcus faecalis in the benzylpenicillin bound form. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Moon, T.M, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2017-12-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
5KSH
 
 | |
6BSQ
 
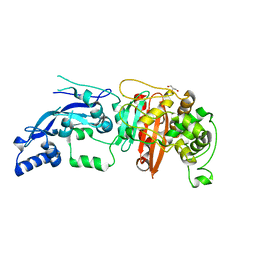 | | Enterococcus faecalis Penicillin Binding Protein 4 (PBP4) | | Descriptor: | CHLORIDE ION, GLYCEROL, PBP4 protein | | Authors: | Moon, T.M, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2017-12-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6G9S
 
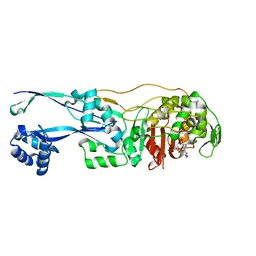 | | Structural basis for the inhibition of E. coli PBP2 | | Descriptor: | (3~{R},6~{S})-6-(aminomethyl)-4-(1,3-oxazol-5-yl)-3-(sulfooxyamino)-3,6-dihydro-2~{H}-pyridine-1-carboxylic acid, Peptidoglycan D,D-transpeptidase MrdA | | Authors: | Ruff, M, Levy, N. | | Deposit date: | 2018-04-11 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis for E. coli Penicillin Binding Protein (PBP) 2 Inhibition, a Platform for Drug Design.
J.Med.Chem., 62, 2019
|
|
8BH1
 
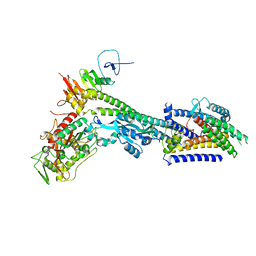 | | Core divisome complex FtsWIQBL from Pseudomonas aeruginosa | | Descriptor: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ, ... | | Authors: | Kaeshammer, L, van den Ent, F, Jeffery, M, Lowe, J. | | Deposit date: | 2022-10-28 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the bacterial divisome core complex and antibiotic target FtsWIQBL.
Nat Microbiol, 8, 2023
|
|
4MNR
 
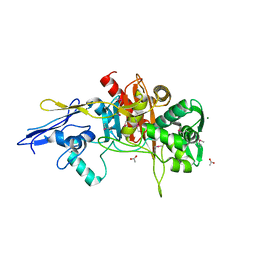 | | Crystal Structure of D,D-Transpeptidase Domain of Peptidoglycan Glycosyltransferase from Eggerthella lenta | | Descriptor: | ACETIC ACID, MAGNESIUM ION, Peptidoglycan glycosyltransferase | | Authors: | Kim, Y, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-11 | | Release date: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Crystal Structure of D,D-Transpeptidase Domain of Peptidoglycan Glycosyltransferase from Eggerthella lenta
To be Published
|
|
8C5W
 
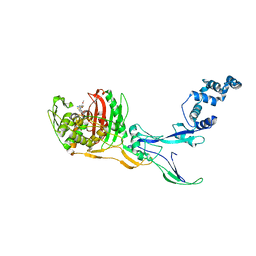 | | Crystal Structure of Penicillin-binding Protein 3 (PBP3) from Staphylococcus Epidermidis in complex with Cefotaxime | | Descriptor: | CEFOTAXIME, C3' cleaved, open, ... | | Authors: | Schwinzer, M, Brognaro, H, Rohde, H, Betzel, C. | | Deposit date: | 2023-01-10 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure and Dynamics of the Penicillin-Binding Protein 3 from Staphylococcus Epidermidis Native and in Complex with Cefotaxime and Vaborbactam
Int J Appl Biol Pharm, 2023
|
|
8C5B
 
 | |
8C5O
 
 | |
4OON
 
 | | Crystal structure of PBP1a in complex with compound 17 ((4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid) | | Descriptor: | (4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid, Penicillin-binding protein 1A | | Authors: | Han, S, Caspers, N, Knafels, J.D. | | Deposit date: | 2014-02-03 | | Release date: | 2014-05-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Siderophore receptor-mediated uptake of lactivicin analogues in gram-negative bacteria.
J.Med.Chem., 57, 2014
|
|
