2J05
 
 | |
2IIM
 
 | | SH3 Domain of Human Lck | | Descriptor: | CALCIUM ION, Proto-oncogene tyrosine-protein kinase LCK, TETRAETHYLENE GLYCOL, ... | | Authors: | Romir, J, Egerer-Sieber, C, Muller, Y.A. | | Deposit date: | 2006-09-28 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structure analysis and solution studies of human Lck-SH3; zinc-induced homodimerization competes with the binding of proline-rich motifs.
J.Mol.Biol., 365, 2007
|
|
2F2W
 
 | | alpha-spectrin SH3 domain R21A mutant | | Descriptor: | SULFATE ION, Spectrin alpha chain, brain | | Authors: | Camara-Artigas, A, Conejero-Lara, F, Casares, S, Lopez-Mayorga, O, Vega, C. | | Deposit date: | 2005-11-18 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cooperative propagation of local stability changes from low-stability and high-stability regions in a SH3 domain
Proteins, 67, 2007
|
|
2EW3
 
 | |
2JM8
 
 | | R21A Spc-SH3 free | | Descriptor: | Spectrin alpha chain, brain | | Authors: | van Nuland, N.A.J, Casares, S, Ab, E, Eshuis, H, Lopez-Mayorga, O, Conejero-Lara, F. | | Deposit date: | 2006-10-25 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The high-resolution NMR structure of the R21A Spc-SH3:P41 complex: Understanding the determinants of binding affinity by comparison with Abl-SH3
Bmc Struct.Biol., 7, 2007
|
|
2K3B
 
 | |
2JW4
 
 | | NMR solution structure of the N-terminal SH3 domain of human Nckalpha | | Descriptor: | Cytoplasmic protein NCK1 | | Authors: | Santiveri, C.M, Borroto, A, Simon, L, Rico, M, Ortiz, A.R, Alarcon, B, Jimenez, M. | | Deposit date: | 2007-10-05 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Interaction between the N-terminal SH3 domain of Nckalpha and CD3epsilon-derived peptides: Non-canonical and canonical recognition motifs
BIOCHEM.BIOPHYS.ACTA PROTEINS & PROTEOMICS, 1794, 2009
|
|
2JTE
 
 | | Third SH3 domain of CD2AP | | Descriptor: | CD2-associated protein | | Authors: | van Nuland, N.A.J, Ortega, R.J, Romero Romero, M, Ab, E, Ora, A, Lopez, M.O, Azuaga, A.I. | | Deposit date: | 2007-07-28 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The high resolution NMR structure of the third SH3 domain of CD2AP.
J.Biomol.Nmr, 39, 2007
|
|
2JXB
 
 | | Structure of CD3epsilon-Nck2 first SH3 domain complex | | Descriptor: | T-cell surface glycoprotein CD3 epsilon chain, Cytoplasmic protein NCK2 | | Authors: | Takeuchi, K, Yang, H, Ng, E, Park, S, Sun, Z.J, Reinherz, E.L, Wagner, G. | | Deposit date: | 2007-11-09 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and functional evidence that Nck interaction with CD3epsilon regulates T-cell receptor activity.
J.Mol.Biol., 380, 2008
|
|
2JMA
 
 | | R21A Spc-SH3:P41 complex | | Descriptor: | P41 peptide, Spectrin alpha chain, brain | | Authors: | van Nuland, N.A.J, Casares, S, Ab, E, Eshuis, H, Lopez-Mayorga, O, Conejero-Lara, F. | | Deposit date: | 2006-10-25 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The high-resolution NMR structure of the R21A Spc-SH3:P41 complex: Understanding the determinants of binding affinity by comparison with Abl-SH3
Bmc Struct.Biol., 7, 2007
|
|
2JS2
 
 | |
2JS0
 
 | |
1PRM
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND PLR1 (AFAPPLPRR) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1PRL
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND PLR1 (AFAPPLPRR) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1UUE
 
 | | a-SPECTRIN SH3 DOMAIN (V44T, D48G MUTANT) | | Descriptor: | SPECTRIN ALPHA CHAIN | | Authors: | Vega, M.C, Fernandez, A, Wilmanns, M, Serrano, L. | | Deposit date: | 2003-12-18 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Solvation in Protein Folding Analysis: Combination of Theoretical and Experimental Approaches
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1QKX
 
 | |
1QKW
 
 | | Alpha-spectrin Src Homology 3 domain, N47G mutant in the distal loop. | | Descriptor: | ALPHA II SPECTRIN, GLYCEROL, SULFATE ION | | Authors: | Vega, M.C, Martinez, J, Serrano, L. | | Deposit date: | 1999-08-16 | | Release date: | 2000-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamic and structural characterization of Asn and Ala residues in the disallowed II' region of the Ramachandran plot.
Protein Sci., 9, 2000
|
|
1WXB
 
 | | Solution structure of the SH3 domain from human epidermal growth factor receptor pathway substrate 8-like protein | | Descriptor: | Epidermal growth factor receptor pathway substrate 8-like protein | | Authors: | Chikayama, E, Kigawa, T, Muto, Y, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-20 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain from human epidermal growth factor receptor pathway substrate 8-like protein
To be Published
|
|
1WLP
 
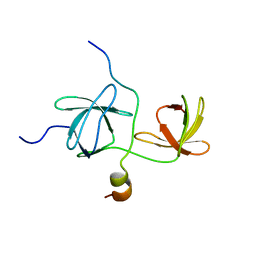 | | Solution Structure Of The P22Phox-P47Phox Complex | | Descriptor: | Cytochrome b-245 light chain, Neutrophil cytosol factor 1 | | Authors: | Ogura, K, Torikai, S, Saikawa, K, Yuzawa, S, Sumimoto, H, Inagaki, F. | | Deposit date: | 2004-06-29 | | Release date: | 2005-10-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the tandem Src homology 3 domains of p47phox complexed with a p22phox-derived proline-rich peptide
J.Biol.Chem., 281, 2006
|
|
1X2P
 
 | | Solution structure of the SH3 domain of the Protein arginine N-methyltransferase 2 | | Descriptor: | Protein arginine N-methyltransferase 2 | | Authors: | Chikayama, E, Kigawa, T, Saito, K, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-04-26 | | Release date: | 2005-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of the Protein arginine N-methyltransferase 2
To be Published
|
|
1WDX
 
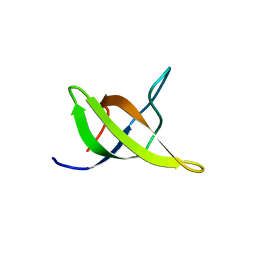 | | Yeast BBC1 SH3 domain, triclinic crystal form | | Descriptor: | Myosin tail region-interacting protein MTI1 | | Authors: | Wilmanns, M, Consani Textor, L, Kursula, P, Kursula, I, Lehmann, F, Song, Y.H. | | Deposit date: | 2004-05-19 | | Release date: | 2005-05-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Yeast BBC1 SH3 domain, triclinic crystal form
To be Published
|
|
1PWT
 
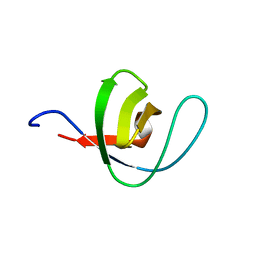 | | THERMODYNAMIC ANALYSIS OF ALPHA-SPECTRIN SH3 AND TWO OF ITS CIRCULAR PERMUTANTS WITH DIFFERENT LOOP LENGTHS: DISCERNING THE REASONS FOR RAPID FOLDING IN PROTEINS | | Descriptor: | ALPHA SPECTRIN | | Authors: | Martinez, J.C, Viguera, A.R, Berisio, R, Wilmanns, M, Mateo, P.L, Filmonov, V.V, Serrano, L. | | Deposit date: | 1998-10-06 | | Release date: | 1999-05-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Thermodynamic analysis of alpha-spectrin SH3 and two of its circular permutants with different loop lengths: discerning the reasons for rapid folding in proteins.
Biochemistry, 38, 1999
|
|
1WYX
 
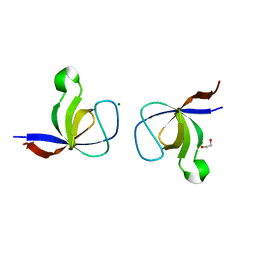 | | The Crystal Structure of the p130Cas SH3 Domain at 1.1 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, CRK-associated substrate, MAGNESIUM ION | | Authors: | Wisniewska, M, Bossenmaier, B, Georges, G, Hesse, F, Dangl, M, Kuenkele, K.P, Ioannidis, I, Huber, R, Engh, R.A. | | Deposit date: | 2005-02-17 | | Release date: | 2005-04-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | The 1.1 A resolution crystal structure of the p130cas SH3 domain and ramifications for ligand selectivity
J.Mol.Biol., 347, 2005
|
|
1X2Q
 
 | | Solution structure of the SH3 domain of the Signal transducing adaptor molecule 2 | | Descriptor: | Signal transducing adapter molecule 2 | | Authors: | Chikayama, E, Kigawa, T, Sato, M, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-04-26 | | Release date: | 2005-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of the Signal transducing adaptor molecule 2
To be Published
|
|
1WX6
 
 | | Solution structure of the SH3 domain of the human cytoplasmic protein NCK2 | | Descriptor: | Cytoplasmic protein NCK2 | | Authors: | Ohnishi, S, Kigawa, T, Tomizawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-20 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of the human cytoplasmic protein NCK2
To be Published
|
|
