3JVY
 
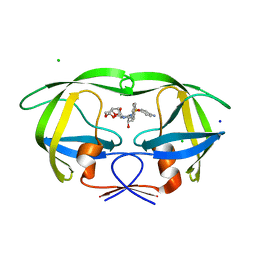 | | HIV-1 Protease Mutant G86A with DARUNAVIR | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, Gag-Pol polyprotein, ... | | Authors: | Tie, Y, Weber, I.T. | | Deposit date: | 2009-09-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Highly conserved glycine 86 and arginine 87 residues contribute differently to the structure and activity of the mature HIV-1 protease
Proteins, 78, 2009
|
|
3QPJ
 
 | |
3QRO
 
 | | HIV-1 protease (mutant Q7K L33I L63I) in complex with a three-armed pyrrolidine-based inhibitor | | Descriptor: | 4-({(3S,4S)-4-[(3,5-dihydroxybenzyl)amino]pyrrolidin-3-yl}[4-(trifluoromethyl)benzyl]sulfamoyl)benzamide, CHLORIDE ION, DITHIANE DIOL, ... | | Authors: | Lindemann, I, Heine, A, Klebe, G. | | Deposit date: | 2011-02-18 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.616 Å) | | Cite: | Design of a series of novel three-armed pyrrolidine-based inhibitors for HIV-1 protease
To be Published
|
|
5DGW
 
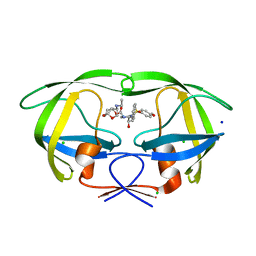 | | Crystal Structure of HIV-1 Protease Inhibitor GRL-105-11A Containing Substituted fused-Tetrahydropyranyl Tetrahydrofuran as P2-Ligand | | Descriptor: | (3R,3aS,4S,7aS)-3-(ethylamino)hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Pol protein, ... | | Authors: | Agniswamy, J, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2015-08-28 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of HIV-1 protease inhibitors containing substituted fused-tetrahydropyranyl tetrahydrofuran as P2-ligands.
Org.Biomol.Chem., 13, 2015
|
|
8DCI
 
 | |
3UF3
 
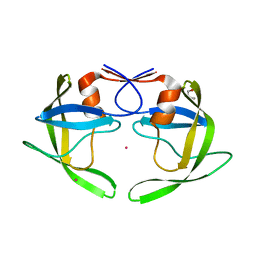 | | Crystal Structure of Multidrug Resistant HIV-1 Protease Clinical isolate PR20 | | Descriptor: | GLYCEROL, HIV-1 protease, YTTRIUM ION | | Authors: | Agniswamy, J, Chen-Hsiang, S, Aniana, A, Sayer, J.M, Louis, J.M, Weber, I.T. | | Deposit date: | 2011-10-31 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | HIV-1 protease with 20 mutations exhibits extreme resistance to clinical inhibitors through coordinated structural rearrangements.
Biochemistry, 51, 2012
|
|
7DPQ
 
 | | HIV-1 Protease D30N mutant | | Descriptor: | Protease | | Authors: | Bihani, S.C, Hosur, M.V. | | Deposit date: | 2020-12-21 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular basis for reduced cleavage activity and drug resistance in D30N HIV-1 protease.
J.Biomol.Struct.Dyn., 2021
|
|
1ZPK
 
 | | Crystal structure of the complex of mutant HIV-1 protease (A71V, V82T, I84V) with a hydroxyethylamine peptidomimetic inhibitor BOC-PHE-PSI[R-CH(OH)CH2NH]-PHE-GLU-PHE-NH2 | | Descriptor: | CHLORIDE ION, N-{(2R,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN, ... | | Authors: | Duskova, J, Skalova, T, Dohnalek, J, Petrokova, H, Hasek, J. | | Deposit date: | 2005-05-17 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mutational Study of Pseudopeptide Inhibitor Binding to HIV-1 Protease; Analysis of Four X-ray Structures
To be Published
|
|
2ZGA
 
 | | HIV-1 protease in complex with a dimethylallyl decorated pyrrolidine based inhibitor (hexagonal space group) | | Descriptor: | (3S,4S),-3,4-Bis-[(4-carbamoyl-benzensulfonyl)-(3-methyl-but-2-enyl)-amino]-pyrrolidine, Protease | | Authors: | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2008-01-21 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Two Solutions for the Same Problem: Multiple Binding Modes of Pyrrolidine-Based HIV-1 Protease Inhibitors
J.Mol.Biol., 410, 2011
|
|
3CKT
 
 | | HIV-1 protease in complex with a dimethylallyl decorated pyrrolidine based inhibitor (orthorombic space group) | | Descriptor: | (3S,4S),-3,4-Bis-[(4-carbamoyl-benzensulfonyl)-(3-methyl-but-2-enyl)-amino]-pyrrolidine, CHLORIDE ION, Protease | | Authors: | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2008-03-17 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Two Solutions for the Same Problem: Multiple Binding Modes of Pyrrolidine-Based HIV-1 Protease Inhibitors
J.Mol.Biol., 410, 2011
|
|
2NPH
 
 | | Crystal structure of HIV1 protease in situ product complex | | Descriptor: | PROTEASE RETROPEPSIN, pentapeptide fragment, tetrapeptide fragment | | Authors: | Hosur, M.V, Das, A, Prashar, V. | | Deposit date: | 2006-10-27 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of HIV-1 protease in situ product complex and observation of a low-barrier hydrogen bond between catalytic aspartates
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3KT2
 
 | | Crystal Structure of N88D mutant HIV-1 Protease | | Descriptor: | Protease | | Authors: | Bihani, S.C, Das, A, Prashar, V, Ferrer, J.L, Hosur, M.V. | | Deposit date: | 2009-11-24 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Resistance mechanism revealed by crystal structures of unliganded nelfinavir-resistant HIV-1 protease non-active site mutants N88D and N88S.
Biochem.Biophys.Res.Commun., 389, 2009
|
|
6P9A
 
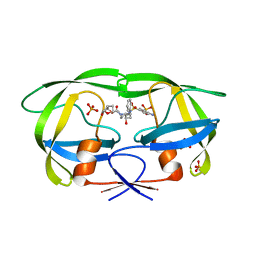 | | HIV-1 Protease multiple mutant PRS5B with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 Protease, PHOSPHATE ION | | Authors: | Kneller, D.W, Agniswamy, J, Weber, I.T. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Highly drug-resistant HIV-1 protease reveals decreased intra-subunit interactions due to clusters of mutations.
Febs J., 287, 2020
|
|
3KDB
 
 | | Crystal Structure of HIV-1 Protease (Q7K, L33I, L63I) in Complex with KNI-10006 | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Protease | | Authors: | Chufan, E.E, Lafont, V, Freire, E, Amzel, L.M. | | Deposit date: | 2009-10-22 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | How much binding affinity can be gained by filling a cavity?
Chem.Biol.Drug Des., 75, 2010
|
|
2WHH
 
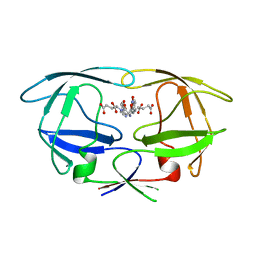 | | HIV-1 protease tethered dimer Q-product complex along with nucleophilic water molecule | | Descriptor: | GLUTAMIC ACID, PARA-NITROPHENYLALANINE, POL PROTEIN | | Authors: | Prashar, V, Bihani, S, Das, A, Ferrer, J.L, Hosur, M.V. | | Deposit date: | 2009-05-05 | | Release date: | 2009-12-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Catalytic Water Co-Existing with a Product Peptide in the Active Site of HIV-1 Protease Revealed by X- Ray Structure Analysis.
Plos One, 4, 2009
|
|
3QIN
 
 | |
5XOS
 
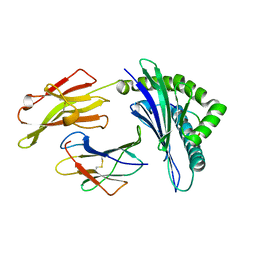 | | Crystal structure of HLA-B35 in complex with a pepetide antigen | | Descriptor: | An HIV reverse transcriptase epitope, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Conserved V delta 1 Binding Geometry in a Setting of Locus-Disparate pHLA Recognition by delta / alpha beta T Cell Receptors (TCRs): Insight into Recognition of HIV Peptides by TCRs.
J. Virol., 91, 2017
|
|
4U7Q
 
 | | Structure of wild-type HIV protease in complex with photosensitive inhibitor PDI-6 | | Descriptor: | N~2~-({[7-(diethylamino)-2-oxo-2H-chromen-4-yl]methoxy}carbonyl)-N-[(2S,4S,5S)-4-hydroxy-1,6-diphenyl-5-{[(1,3-thiazol-5-ylmethoxy)carbonyl]amino}hexan-2-yl]-L-valinamide, V-1 protease | | Authors: | Pachl, P, Rezacova, P, Schimer, J. | | Deposit date: | 2014-07-31 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Triggering HIV polyprotein processing by light using rapid photodegradation of a tight-binding protease inhibitor.
Nat Commun, 6, 2015
|
|
7MAD
 
 | | HIV-1 Protease (I84V) in Complex with PD5 (LR4-22) | | Descriptor: | Protease, diethyl [(4-{(2S,3R)-4-{[(1,3-benzothiazol-6-yl)sulfonyl](2-methylpropyl)amino}-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with PD5 (LR4-22)
To Be Published
|
|
7MAC
 
 | | HIV-1 Protease (I84V) in Complex with PD4 (LR4-23) | | Descriptor: | Protease, diethyl [(4-{(2S,3R)-4-{[(2H-1,3-benzodioxol-5-yl)sulfonyl](2-methylpropyl)amino}-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with PD4 (LR4-23)
To Be Published
|
|
7MAR
 
 | | Drug Resistant HIV-1 Protease (L10F, M46I, I47V, I50V, F53L, L63P, I72V, G73S, V82I, I85V) in Complex with DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Drug Resistant HIV-1 Protease (L10F, M46I, I47V, I50V, F53L, L63P, I72V, G73S, V82I, I85V) in Complex with DRV
To Be Published
|
|
3I8W
 
 | |
1DIF
 
 | | HIV-1 PROTEASE IN COMPLEX WITH A DIFLUOROKETONE CONTAINING INHIBITOR A79285 | | Descriptor: | BETA-MERCAPTOETHANOL, HIV-1 PROTEASE, N-{1-BENZYL-2,2-DIFLUORO-3,3-DIHYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE | | Authors: | Silva, A.M, Cachau, R.E, Sham, H.L, Erickson, J.W. | | Deposit date: | 1995-10-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition and catalytic mechanism of HIV-1 aspartic protease.
J.Mol.Biol., 255, 1996
|
|
3NLS
 
 | | Crystal Structure of HIV-1 Protease in Complex with KNI-10772 | | Descriptor: | (4R)-3-[(2R,3S)-3-{[(2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H- inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Gabelli, S.B, Kawasaki, Y, Freire, E, Amzel, L.M. | | Deposit date: | 2010-06-21 | | Release date: | 2011-09-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of HIV-1 Protease in Complex with KNI-10772
To be Published
|
|
1XL5
 
 | | HIV-1 Protease in complex with amidhyroxysulfone | | Descriptor: | CHLORIDE ION, N-{(1S)-1-(3-BROMOBENZYL)-4-[(4-BROMOPHENYL)SULFONYL]-6-METHYL-2-OXOHEPTYL}-2-(2,6-DIMETHYLPHENOXY)ACETAMIDE, PROTEASE RETROPEPSIN | | Authors: | Boettcher, J, Specker, E, Heine, A, Klebe, G. | | Deposit date: | 2004-09-30 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | An Old Target Revisited: Two New Privileged Skeletons and an Unexpected Binding Mode For HIV-Protease Inhibitors
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
