4CEL
 
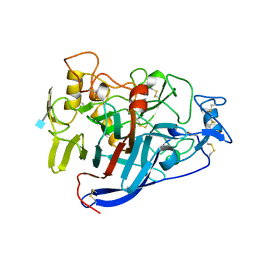 | | ACTIVE-SITE MUTANT D214N DETERMINED AT PH 6.0 WITH NO LIGAND BOUND IN THE ACTIVE SITE | | 分子名称: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION | | 著者 | Divne, C, Stahlberg, J, Jones, T.A. | | 登録日 | 1996-08-24 | | 公開日 | 1997-03-12 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Activity studies and crystal structures of catalytically deficient mutants of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 264, 1996
|
|
1ISQ
 
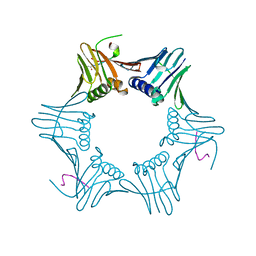 | | Pyrococcus furiosus PCNA complexed with RFCL PIP-box peptide | | 分子名称: | Proliferating Cell Nuclear Antigen, replication factor C large subunit | | 著者 | Matsumiya, S, Ishino, S, Ishino, Y, Morikawa, K. | | 登録日 | 2001-12-19 | | 公開日 | 2002-10-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Physical interaction between proliferating cell nuclear antigen and
replication factor C from Pyrococcus furiosus
Genes Cells, 7, 2002
|
|
1J0D
 
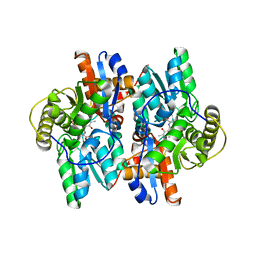 | | ACC deaminase mutant complexed with ACC | | 分子名称: | 1-aminocyclopropane-1-carboxylate deaminase, N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID | | 著者 | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | 登録日 | 2002-11-12 | | 公開日 | 2003-05-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
1IYV
 
 | |
1E3H
 
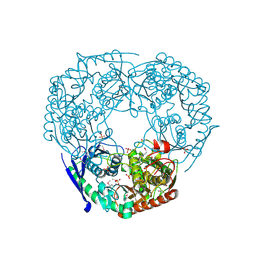 | | SeMet derivative of Streptomyces antibioticus PNPase/GPSI enzyme | | 分子名称: | GUANOSINE PENTAPHOSPHATE SYNTHETASE, SULFATE ION | | 著者 | Symmons, M.F, Jones, G.H, Luisi, B.F. | | 登録日 | 2000-06-15 | | 公開日 | 2000-11-05 | | 最終更新日 | 2019-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A Duplicated Fold is the Structural Basis for Polynucleotide Phosphorylase Catalytic Activity, Processivity, and Regulation
Structure, 8, 2000
|
|
2DKV
 
 | | Crystal structure of class I chitinase from Oryza sativa L. japonica | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, chitinase | | 著者 | Kezuka, Y, Nishizawa, Y, Watanabe, T, Nonaka, T. | | 登録日 | 2006-04-14 | | 公開日 | 2007-05-01 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of full-length class I chitinase from rice revealed by X-ray crystallography and small-angle X-ray scattering.
Proteins, 78, 2010
|
|
1XNA
 
 | |
1XNT
 
 | |
1IVF
 
 | |
1CQ4
 
 | |
2DRZ
 
 | |
1II5
 
 | |
4CCH
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in an open binary complex with d5SICS as templating nucleotide | | 分子名称: | 5'-D(*AP*AP*CP*LHOP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP* C)-3', 5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*DOC)-3', DNA POLYMERASE I, ... | | 著者 | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | 登録日 | 2013-10-23 | | 公開日 | 2013-12-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
2LPR
 
 | |
2DQT
 
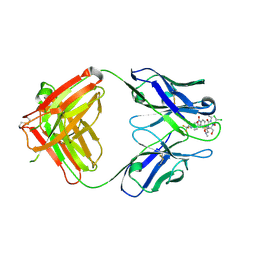 | | High resolution crystal structure of the complex of the hydrolytic antibody Fab 6D9 and a transition-state analog | | 分子名称: | IMMUNOGLOBULIN 6D9, [1-(3-DIMETHYLAMINO-PROPYL)-3-ETHYL-UREIDO]-[4-(2,2,2-TRIFLUORO-ACETYLAMINO)-BENZYL]PHOSPHINIC ACID-2-(2,2-DIHYDRO-ACETYLAMINO)-3-HYDROXY-1-(4-NITROPHENYL)-PROPYL ESTER | | 著者 | Kristensen, O, Vassylyev, D.G, Tanaka, F, Ito, N, Morikawa, K, Fujii, I. | | 登録日 | 2006-05-30 | | 公開日 | 2006-06-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Thermodynamic and structural basis for transition-state stabilization in antibody-catalyzed hydrolysis
J.Mol.Biol., 369, 2007
|
|
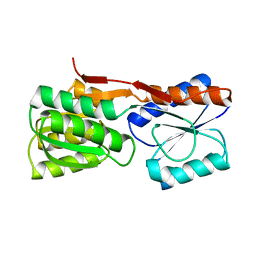
2DRI
 
 | |
1DT0
 
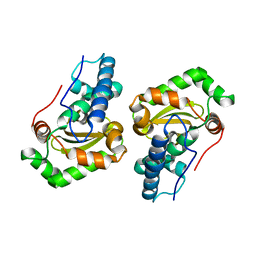 | | CLONING, SEQUENCE, AND CRYSTALLOGRAPHIC STRUCTURE OF RECOMBINANT IRON SUPEROXIDE DISMUTASE FROM PSEUDOMONAS OVALIS | | 分子名称: | FE (III) ION, SUPEROXIDE DISMUTASE | | 著者 | Bond, C.J, Huang, J, Hajduk, R, Flick, K, Heath, P, Stoddard, B.L. | | 登録日 | 2000-01-10 | | 公開日 | 2000-12-20 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Cloning, sequence and crystallographic structure of recombinant iron superoxide dismutase from Pseudomonas ovalis.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1DQO
 
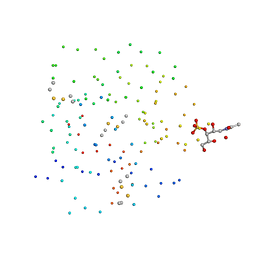 | | Crystal structure of the cysteine rich domain of mannose receptor complexed with Acetylgalactosamine-4-sulfate | | 分子名称: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, MANNOSE RECEPTOR | | 著者 | Liu, Y, Chirino, A.J, Misulovin, Z, Leteux, C, Feizi, T, Nussenzweig, M.C, Bjorkman, P.J. | | 登録日 | 2000-01-04 | | 公開日 | 2000-05-10 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of the cysteine-rich domain of mannose receptor complexed with a sulfated carbohydrate ligand.
J.Exp.Med., 191, 2000
|
|
2R0K
 
 | | Protease domain of HGFA with inhibitor Fab58 | | 分子名称: | Hepatocyte growth factor activator, antibody heavy chain of Fab58, Fab portion only, ... | | 著者 | Eigenbrot, C, Shia, S. | | 登録日 | 2007-08-20 | | 公開日 | 2007-12-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.51 Å) | | 主引用文献 | Structural insight into distinct mechanisms of protease inhibition by antibodies.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2ALA
 
 | | Crystal structure of the Semliki Forest Virus envelope protein E1 in its monomeric conformation. | | 分子名称: | Structural polyprotein (P130) | | 著者 | Roussel, A, Lescar, J, Vaney, M.C, Wengler, G, Wengler, G, Rey, F.A. | | 登録日 | 2005-08-05 | | 公開日 | 2006-01-17 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure and interactions at the viral surface of the envelope protein E1 of Semliki Forest virus.
Structure, 14, 2006
|
|
1E3P
 
 | | tungstate derivative of Streptomyces antibioticus PNPase/GPSI enzyme | | 分子名称: | Polyribonucleotide nucleotidyltransferase, SULFATE ION, TUNGSTATE(VI)ION | | 著者 | Symmons, M.F, Jones, G.H, Luisi, B.F. | | 登録日 | 2000-06-20 | | 公開日 | 2000-11-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A Duplicated Fold is the Structural Basis for Polynucleotide Phosphorylase Catalytic Activity, Processivity, and Regulation
Structure, 8, 2000
|
|
1DTK
 
 | |
1AM2
 
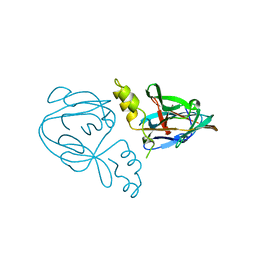 | |
1NIY
 
 | |
2CL5
 
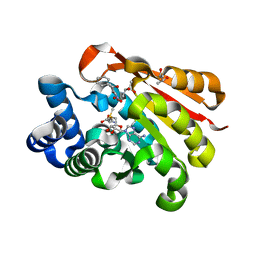 | | Catechol-O-methyltransferase in complex with an inhibitor | | 分子名称: | (3,4-DIHYDROXY-2-NITROPHENYL)(PHENYL)METHANONE, (R,R)-2,3-BUTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Palma, P.N, Rodrigues, M.L, Archer, M, Bonifacio, M.J, Loureiro, A.I, Learmonth, D.A, Carrondo, M.A, Soares-Da-Silva, P. | | 登録日 | 2006-04-26 | | 公開日 | 2006-06-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Comparative Study of Ortho- and Meta-Nitrated Inhibitors of Catechol-O-Methyltransferase: Interactions with the Active Site and Regioselectivity of O-Methylation.
Mol.Pharmacol., 70, 2006
|
|
