6H1B
 
 | | Structure of amide bond synthetase Mcba K483A mutant from Marinactinospora thermotolerans | | 分子名称: | 1-ethanoyl-9~{H}-pyrido[3,4-b]indole-3-carboxylic acid, ADENOSINE MONOPHOSPHATE, Fatty acid CoA ligase | | 著者 | Rowlinson, B, Petchey, M, Cuetos, A, Frese, A, Dannevald, S, Grogan, G. | | 登録日 | 2018-07-11 | | 公開日 | 2018-09-05 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The Broad Aryl Acid Specificity of the Amide Bond Synthetase McbA Suggests Potential for the Biocatalytic Synthesis of Amides.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6B5B
 
 | | Cryo-EM structure of the NAIP5-NLRC4-flagellin inflammasome | | 分子名称: | Baculoviral IAP repeat-containing protein 1e, Flagellin, NLR family CARD domain-containing protein 4 | | 著者 | Tenthorey, J.L, Haloupek, N, Lopez-Blanco, J.R, Grob, P, Adamson, E, Hartenian, E, Lind, N.A, Bourgeois, N.M, Chacon, P, Nogales, E, Vance, R.E. | | 登録日 | 2017-09-29 | | 公開日 | 2017-11-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (5.2 Å) | | 主引用文献 | The structural basis of flagellin detection by NAIP5: A strategy to limit pathogen immune evasion.
Science, 358, 2017
|
|
5FJO
 
 | |
4L6E
 
 | | Crystal Structure of the RanBD1 fourth domain of E3 SUMO-protein ligase RanBP2. Northeast Structural Genomics Consortium (NESG) Target HR9193b | | 分子名称: | E3 SUMO-protein ligase RanBP2 | | 著者 | Vorobiev, S, Su, M, Seetharaman, J, Mao, L, Xiao, R, Maglaqui, M, Kogan, S, Wang, H, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-06-12 | | 公開日 | 2013-06-26 | | 実験手法 | X-RAY DIFFRACTION (2.496 Å) | | 主引用文献 | Crystal Structure of the RanBD1 fourth domain of E3 SUMO-protein ligase RanBP2.
To be Published
|
|
5A4W
 
 | | AtGSTF2 from Arabidopsis thaliana in complex with quercetrin | | 分子名称: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4-oxo-4H-chromen-3-yl 6-deoxy-alpha-L-mannopyranoside, ACETATE ION, GLUTATHIONE S-TRANSFERASE F2 | | 著者 | Ahmad, L, Rylott, E, Bruce, N.C, Edwards, R, Grogan, G. | | 登録日 | 2015-06-15 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural evidence for Arabidopsis glutathione transferase AtGSTF2 functioning as a transporter of small organic ligands.
FEBS Open Bio, 7, 2017
|
|
5A9T
 
 | | Imine Reductase from Amycolatopsis orientalis in complex with (R)- Methyltetrahydroisoquinoline | | 分子名称: | (1R)-1-methyl-1,2,3,4-tetrahydroisoquinoline, ACETATE ION, CALCIUM ION, ... | | 著者 | Man, H, Aleku, G, Turner, N.J, Grogan, G. | | 登録日 | 2015-07-22 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Stereoselectivity and Structural Characterization of an Imine Reductase (Ired) from Amycolatopsis Orientalis
Acs Catalysis, 6, 2016
|
|
4LLJ
 
 | | Crystal structure of PDE10A2 with fragment ZT214 | | 分子名称: | 2H-isoindole-1,3-diamine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | 著者 | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | 登録日 | 2013-07-09 | | 公開日 | 2014-02-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LM0
 
 | | Crystal structure of PDE10A2 with fragment ZT448 | | 分子名称: | 5-NITROINDAZOLE, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | 著者 | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | 登録日 | 2013-07-09 | | 公開日 | 2014-02-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
7DXI
 
 | |
5FJR
 
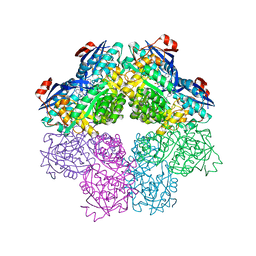 | |
5FR9
 
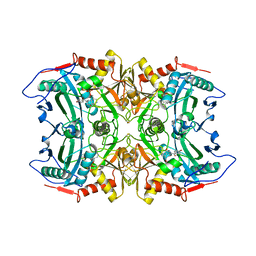 | | Structure of transaminase ATA-117 arRmut11 from Arthrobacter sp. KNK168 inhibited with 1-(4-Bromophenyl)-2-fluoroethylamine | | 分子名称: | (R)-AMINE TRANSAMINASE, [4-[3-(4-bromophenyl)-3-oxidanylidene-propyl]-6-methyl-5-oxidanyl-pyridin-3-yl]methyl phosphate | | 著者 | Cuetos, A, Kroutil, W, Lavandera, I, Grogan, G. | | 登録日 | 2015-12-16 | | 公開日 | 2016-03-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Catalytic Promiscuity of Transaminases: Preparation of Enantioenriched Beta-Fluoroamines by Formal Tandem Hydrodefluorination/Deamination.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
4LLK
 
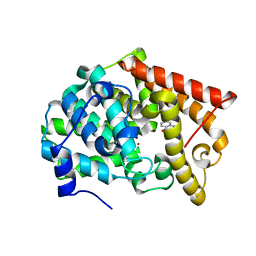 | | Crystal structure of PDE10A2 with fragment ZT217 | | 分子名称: | 2-methylquinazolin-4(3H)-one, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | 著者 | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | 登録日 | 2013-07-09 | | 公開日 | 2014-02-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
6GVC
 
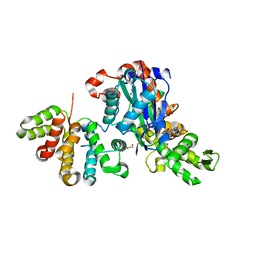 | | Structure of ArhGAP12 bound to G-Actin | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | 著者 | Mouilleron, S, Treisman, R, Diring, J. | | 登録日 | 2018-06-20 | | 公開日 | 2019-03-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | RPEL-family rhoGAPs link Rac/Cdc42 GTP loading to G-actin availability.
Nat.Cell Biol., 21, 2019
|
|
5FJP
 
 | |
5FWN
 
 | | Imine Reductase from Amycolatopsis orientalis. Closed form in in complex with (R)- Methyltetrahydroisoquinoline | | 分子名称: | (1R)-1-methyl-1,2,3,4-tetrahydroisoquinoline, IMINE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Man, H, Aleku, G, Turner, N.J, Grogan, G. | | 登録日 | 2016-02-18 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Stereoselectivity and Structural Characterization of an Imine Reductase (Ired) from Amycolatopsis Orientalis
Acs Catalysis, 6, 2016
|
|
6I04
 
 | | Crystal structure of Sema domain of the Met receptor in complex with FAB | | 分子名称: | Fab heavy chain, Fab light chain, Hepatocyte growth factor receptor | | 著者 | Casaletto, J.B, Geddie, M.L, Abu-Yousif, A.O, Masson, K, Fulgham, A, Boudot, A, Maiwald, T, Kearns, J.D, Kohli, N, Su, S, Razlog, M, Raue, A, Kalra, A, Hakansson, M, Logan, D.T, Welin, M, Chattopadhyay, S, Harms, B.D, Nielsen, U.B, Schoeberl, B, Lugovskoy, A.A, MacBeath, G. | | 登録日 | 2018-10-25 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | MM-131, a bispecific anti-Met/EpCAM mAb, inhibits HGF-dependent and HGF-independent Met signaling through concurrent binding to EpCAM.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4XCR
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form, mutant I35A | | 分子名称: | Superoxide dismutase [Cu-Zn] | | 著者 | Wang, H, Logan, D.T, Danielsson, J, Mu, X, Binolfi, A, Theillet, F, Bekei, B, Lang, L, Wennerstrom, H, Selenko, P, Oliveberg, M. | | 登録日 | 2014-12-18 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.602 Å) | | 主引用文献 | Thermodynamics of protein destabilization in live cells.
Proc. Natl. Acad. Sci. U.S.A., 112, 2015
|
|
6HF2
 
 | | The structure of BoMan26B, a GH26 beta-mannanase from Bacteroides ovatus | | 分子名称: | CALCIUM ION, CHLORIDE ION, Glycosyl hydrolase family 26 | | 著者 | Bagenholm, V, Logan, D.T, Stalbrand, H. | | 登録日 | 2018-08-21 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | A surface-exposed GH26 beta-mannanase fromBacteroides ovatus: Structure, role, and phylogenetic analysis ofBoMan26B.
J.Biol.Chem., 294, 2019
|
|
6HF4
 
 | | The structure of BoMan26B, a GH26 beta-mannanase from Bacteroides ovatus, complexed with G1M4 | | 分子名称: | CALCIUM ION, CHLORIDE ION, Glycosyl hydrolase family 26, ... | | 著者 | Bagenholm, V, Logan, D.T, Stalbrand, H. | | 登録日 | 2018-08-21 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.781 Å) | | 主引用文献 | A surface-exposed GH26 beta-mannanase fromBacteroides ovatus: Structure, role, and phylogenetic analysis ofBoMan26B.
J.Biol.Chem., 294, 2019
|
|
6HGN
 
 | |
4Y04
 
 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) from Porphyromonas gingivalis (Space) | | 分子名称: | GLYCEROL, POTASSIUM ION, Peptidase S46 | | 著者 | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Inaka, K, Tanaka, H, Yamada, M, Ohta, K, Nonaka, T, Ogasawara, W, Tanaka, N. | | 登録日 | 2015-02-05 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity.
Sci Rep, 5, 2015
|
|
4MSH
 
 | | Crystal Structure of PDE10A2 with fragment ZT0143 ((2S)-4-chloro-2,3-dihydro-1,3-benzothiazol-2-amine) | | 分子名称: | 4-chloro-1,3-benzothiazol-2-amine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | 著者 | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | 登録日 | 2013-09-18 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
5FJU
 
 | |
4L1L
 
 | | Rat PKC C2 domain bound to CD | | 分子名称: | CADMIUM ION, Protein kinase C alpha type, SULFATE ION | | 著者 | Morales, K.M, Yang, Y, Long, Z, Li, P, Taylor, A.B, Hart, P.J, Igumenova, T.I. | | 登録日 | 2013-06-03 | | 公開日 | 2013-08-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Cd(2+) as a ca(2+) surrogate in protein-membrane interactions: isostructural but not isofunctional.
J.Am.Chem.Soc., 135, 2013
|
|
5FJT
 
 | |
