3QTE
 
 | |
5YPB
 
 | | p62/SQSTM1 ZZ domain with His-peptide | | 分子名称: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | 著者 | Kwon, D.H, Kim, L, Song, H.K. | | 登録日 | 2017-11-01 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
5YPH
 
 | | p62/SQSTM1 ZZ domain with Ile-peptide | | 分子名称: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | 著者 | Kwon, D.H, Kim, L, Song, H.K. | | 登録日 | 2017-11-01 | | 公開日 | 2018-08-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.629 Å) | | 主引用文献 | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
5YP8
 
 | | p62/SQSTM1 ZZ domain with Arg-peptide | | 分子名称: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | 著者 | Kwon, D.H, Kim, L, Song, H.K. | | 登録日 | 2017-11-01 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.448 Å) | | 主引用文献 | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
5YPG
 
 | | p62/SQSTM1 ZZ domain with Leu-peptide | | 分子名称: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | 著者 | Kwon, D.H, Kim, L, Song, H.K. | | 登録日 | 2017-11-01 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.199 Å) | | 主引用文献 | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
7U69
 
 | |
7U3M
 
 | |
7U6A
 
 | |
5MT3
 
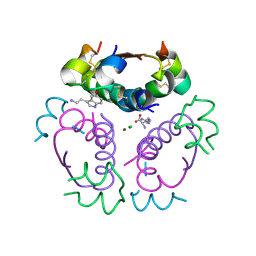 | | Human insulin in complex with serotonin and arginine | | 分子名称: | ARGININE, CHLORIDE ION, Insulin, ... | | 著者 | Brzozowski, A.M, Turkenburg, J.P, Jiracek, J, Zakova, L. | | 登録日 | 2017-01-06 | | 公開日 | 2017-04-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Computational and structural evidence for neurotransmitter-mediated modulation of the oligomeric states of human insulin in storage granules.
J. Biol. Chem., 292, 2017
|
|
5MT9
 
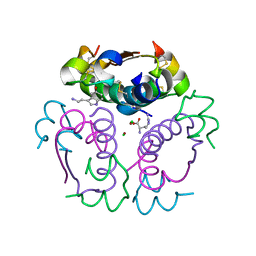 | | Human insulin in complex with serotonin and arginine | | 分子名称: | ARGININE, CHLORIDE ION, Insulin, ... | | 著者 | Brzozowski, A.M, Turkenburg, J.P, Jiracek, J, Zakova, L. | | 登録日 | 2017-01-07 | | 公開日 | 2017-04-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Computational and structural evidence for neurotransmitter-mediated modulation of the oligomeric states of human insulin in storage granules.
J. Biol. Chem., 292, 2017
|
|
7U6B
 
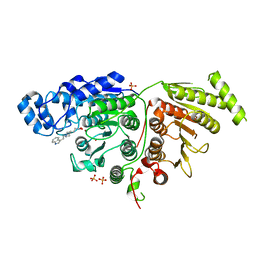 | |
5YPF
 
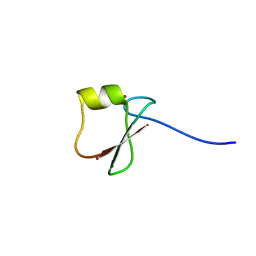 | | p62/SQSTM1 ZZ domain with Trp-peptide | | 分子名称: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | 著者 | Kwon, D.H, Kim, L, Song, H.K. | | 登録日 | 2017-11-01 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.951 Å) | | 主引用文献 | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
5JST
 
 | | MBP fused MDV1 coiled coil | | 分子名称: | ACETATE ION, GLYCEROL, Maltose-binding periplasmic protein,Mitochondrial division protein 1, ... | | 著者 | Kim, B.-W, Song, H.K. | | 登録日 | 2016-05-09 | | 公開日 | 2017-03-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.199 Å) | | 主引用文献 | ACCORD: an assessment tool to determine the orientation of homodimeric coiled-coils.
Sci Rep, 7, 2017
|
|
5NJR
 
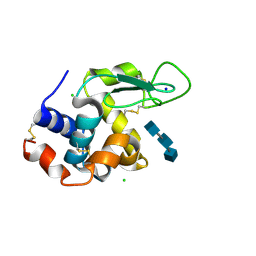 | | Mix-and-diffuse serial synchrotron crystallography: structure of N,N',N''-Triacetylchitotriose bound to Lysozyme with 50s time-delay, phased with 4ET8 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | 著者 | Oberthuer, D, Meents, A, Beyerlein, K.R, Chapman, H.N, Lieseke, J. | | 登録日 | 2017-03-29 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mix-and-diffuse serial synchrotron crystallography.
IUCrJ, 4, 2017
|
|
5YPE
 
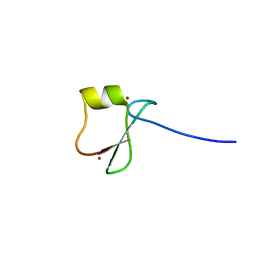 | | p62/SQSTM1 ZZ domain with Tyr-peptide | | 分子名称: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | 著者 | Kwon, D.H, Kim, L, Song, H.K. | | 登録日 | 2017-11-01 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.851 Å) | | 主引用文献 | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
7U59
 
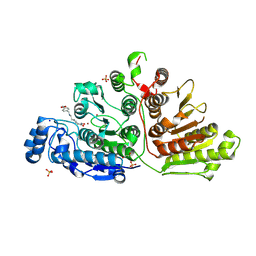 | |
5YPA
 
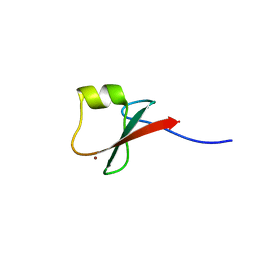 | | p62/SQSTM1 ZZ domain with Lys-peptide | | 分子名称: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | 著者 | Kwon, D.H, Kim, L, Song, H.K. | | 登録日 | 2017-11-01 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
5YP7
 
 | | p62/SQSTM1 ZZ domain | | 分子名称: | Sequestosome-1, ZINC ION | | 著者 | Kwon, D.H, Kim, L, Song, H.K. | | 登録日 | 2017-11-01 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.424 Å) | | 主引用文献 | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
1NE3
 
 | | Solution structure of ribosomal protein S28E from Methanobacterium Thermoautotrophicum. Ontario Centre for Structural Proteomics target MTH0256_1_68; Northeast Structural Genomics Target TT744 | | 分子名称: | 30S ribosomal protein S28E | | 著者 | Wu, B, Pineda-Lucena, A, Yee, A, Cort, J.R, Ramelot, T.A, Kennedy, M, Edwards, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2002-12-10 | | 公開日 | 2003-12-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of ribosomal protein S28E from Methanobacterium thermoautotrophicum.
Protein Sci., 12, 2003
|
|
5JU7
 
 | | DNA BINDING DOMAIN OF E.COLI CADC | | 分子名称: | Transcriptional activator CadC, ZINC ION | | 著者 | Janowski, R, Schlundt, A, Sattler, M, Niessing, D. | | 登録日 | 2016-05-10 | | 公開日 | 2017-04-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure-function analysis of the DNA-binding domain of a transmembrane transcriptional activator.
Sci Rep, 7, 2017
|
|
1OVO
 
 | | CRYSTALLOGRAPHIC REFINEMENT OF JAPANESE QUAIL OVOMUCOID, A KAZAL-TYPE INHIBITOR, AND MODEL BUILDING STUDIES OF COMPLEXES WITH SERINE PROTEASES | | 分子名称: | OVOMUCOID THIRD DOMAIN | | 著者 | Weber, E, Papamokos, E, Bode, W, Huber, R, Kato, I, Laskowskijunior, M. | | 登録日 | 1982-01-18 | | 公開日 | 1982-05-26 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystallographic refinement of Japanese quail ovomucoid, a Kazal-type inhibitor, and model building studies of complexes with serine proteases.
J.Mol.Biol., 158, 1982
|
|
8AXW
 
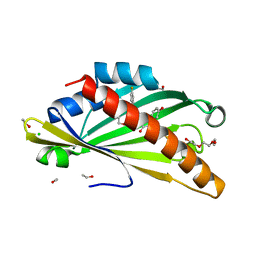 | | The structure of mouse AsterC (GramD1c) with Ezetimibe | | 分子名称: | (3~{R},4~{S})-1-(4-fluorophenyl)-3-[(3~{S})-3-(4-fluorophenyl)-3-oxidanyl-propyl]-4-(4-hydroxyphenyl)azetidin-2-one, CHLORIDE ION, ETHANOL, ... | | 著者 | Fairall, L, Xiao, X, Burger, L, Tontonoz, P, Schwabe, J.W.R. | | 登録日 | 2022-09-01 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Aster-dependent nonvesicular transport facilitates dietary cholesterol uptake.
Science, 382, 2023
|
|
5UIC
 
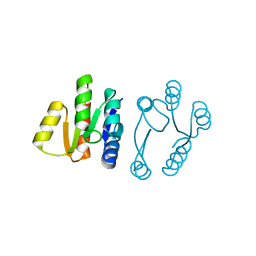 | |
8BAH
 
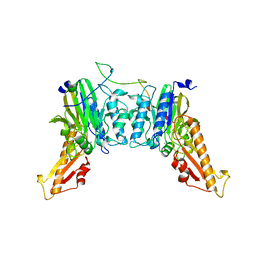 | | Human Mre11-Nbs1 complex | | 分子名称: | Double-strand break repair protein MRE11, MANGANESE (II) ION, Nibrin | | 著者 | Bartho, J.D, Rotheneder, M, Stakyte, K, Lammens, K, Hopfner, K.P. | | 登録日 | 2022-10-11 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (4.13 Å) | | 主引用文献 | Cryo-EM structure of the Mre11-Rad50-Nbs1 complex reveals the molecular mechanism of scaffolding functions.
Mol.Cell, 83, 2023
|
|
5K5M
 
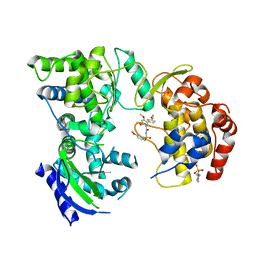 | |
