7TID
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) and primer-template DNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*AP*GP*AP*CP*AP*CP*TP*AP*CP*GP*AP*GP*TP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*AP*TP*GP*TP*AP*CP*TP*CP*GP*TP*AP*GP*TP*GP*TP*CP*T)-3'), ... | | 著者 | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | 登録日 | 2022-01-13 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TIB
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) and primer-template DNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*AP*GP*AP*CP*AP*CP*TP*AP*CP*GP*AP*GP*TP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*AP*TP*GP*TP*AP*CP*TP*CP*GP*TP*AP*GP*TP*GP*TP*CP*T)-3'), ... | | 著者 | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | 登録日 | 2022-01-13 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7THJ
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | 登録日 | 2022-01-11 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7THV
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | 登録日 | 2022-01-12 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7PX9
 
 | | Substrate-engaged mycobacterial Proteasome-associated ATPase - focused 3D refinement (state A) | | 分子名称: | AAA ATPase forming ring-shaped complexes, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Jomaa, A, Kavalchuk, M, Weber-Ban, E. | | 登録日 | 2021-10-08 | | 公開日 | 2022-01-19 | | 最終更新日 | 2022-01-26 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis of prokaryotic ubiquitin-like protein engagement and translocation by the mycobacterial Mpa-proteasome complex.
Nat Commun, 13, 2022
|
|
7PXA
 
 | |
7PXC
 
 | | Substrate-engaged mycobacterial Proteasome-associated ATPase in complex with open-gate 20S CP - composite map (state A) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Jomaa, A, Kavalchuk, M, Weber-Ban, E. | | 登録日 | 2021-10-08 | | 公開日 | 2022-01-19 | | 最終更新日 | 2022-01-26 | | 実験手法 | ELECTRON MICROSCOPY (3.84 Å) | | 主引用文献 | Structural basis of prokaryotic ubiquitin-like protein engagement and translocation by the mycobacterial Mpa-proteasome complex.
Nat Commun, 13, 2022
|
|
7PXB
 
 | | Substrate-engaged mycobacterial Proteasome-associated ATPase - focused 3D refinement (state B) | | 分子名称: | AAA ATPase forming ring-shaped complexes, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Jomaa, A, Kavalchuk, M, Weber-Ban, E. | | 登録日 | 2021-10-08 | | 公開日 | 2022-01-19 | | 最終更新日 | 2022-01-26 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural basis of prokaryotic ubiquitin-like protein engagement and translocation by the mycobacterial Mpa-proteasome complex.
Nat Commun, 13, 2022
|
|
7PXD
 
 | | Substrate-engaged mycobacterial Proteasome-associated ATPase in complex with open-gate 20S CP - composite map (state B) | | 分子名称: | AAA ATPase forming ring-shaped complexes, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Jomaa, A, Kavalchuk, M, Weber-Ban, E. | | 登録日 | 2021-10-08 | | 公開日 | 2022-01-19 | | 最終更新日 | 2022-01-26 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural basis of prokaryotic ubiquitin-like protein engagement and translocation by the mycobacterial Mpa-proteasome complex.
Nat Commun, 13, 2022
|
|
7SXO
 
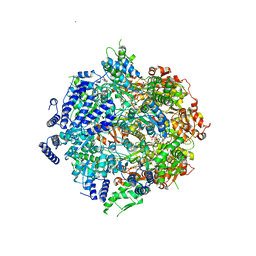 | | Yeast Lon (PIM1) with endogenous substrate | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Lon protease homolog, ... | | 著者 | Yang, J, Song, A.S, Wiseman, R.L, Lander, G.C. | | 登録日 | 2021-11-24 | | 公開日 | 2022-01-12 | | 最終更新日 | 2022-07-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structure of hexameric yeast Lon protease (PIM1) highlights the importance of conserved structural elements.
J.Biol.Chem., 298, 2022
|
|
7OXO
 
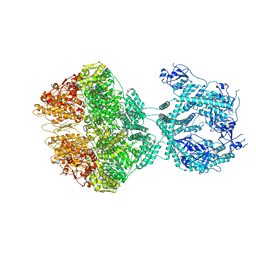 | | human LonP1, R-state, incubated in AMPPCP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | 著者 | Abrahams, J.P, Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T. | | 登録日 | 2021-06-22 | | 公開日 | 2021-12-22 | | 最終更新日 | 2022-11-09 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
7FID
 
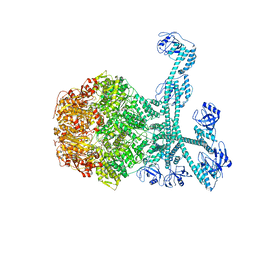 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon proteasecomplex (conformation 1) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | 登録日 | 2021-07-31 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.44 Å) | | 主引用文献 | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7FIZ
 
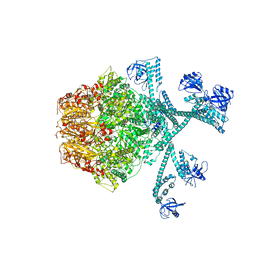 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex (conformation 3) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | 登録日 | 2021-08-01 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7FIE
 
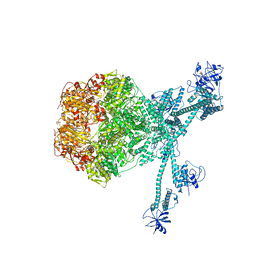 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex (conformation 2) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | 登録日 | 2021-07-31 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.36 Å) | | 主引用文献 | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7FD4
 
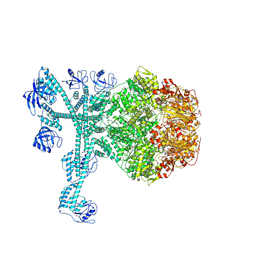 | | A complete three-dimensional structure of the Lon protease translocating a protein substrate (conformation 1) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Lon protease, ... | | 著者 | Li, S, Hsieh, K, Kuo, C, Lee, S, Pintilie, G, Zhang, K, Chang, C. | | 登録日 | 2021-07-16 | | 公開日 | 2021-11-03 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Complete three-dimensional structures of the Lon protease translocating a protein substrate.
Sci Adv, 7, 2021
|
|
7FD5
 
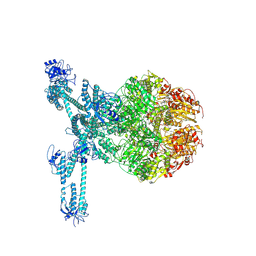 | | A complete three-dimensional structure of the Lon protease translocating a protein substrate (conformation 2) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Lon protease, ... | | 著者 | Li, S, Hsieh, K, Kuo, C, Lee, S, Pintilie, G, Zhang, K, Chang, C. | | 登録日 | 2021-07-16 | | 公開日 | 2021-11-03 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Complete three-dimensional structures of the Lon protease translocating a protein substrate.
Sci Adv, 7, 2021
|
|
7P6U
 
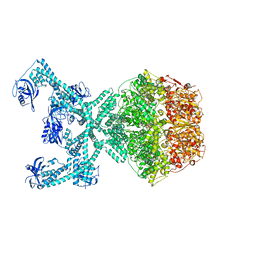 | | Lon protease from Thermus Thermophilus | | 分子名称: | (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK), Lon protease, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Coscia, F, Lowe, J. | | 登録日 | 2021-07-18 | | 公開日 | 2021-10-27 | | 最終更新日 | 2021-11-17 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structure of the full-length Lon protease from Thermus thermophilus.
Febs Lett., 595, 2021
|
|
7K59
 
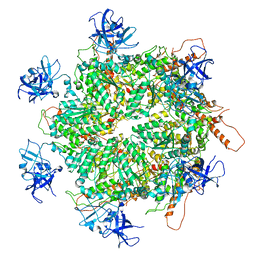 | | Structure of apo VCP hexamer generated from bacterially recombinant VCP/p97 | | 分子名称: | Transitional endoplasmic reticulum ATPase | | 著者 | Yu, G, Bai, Y, Li, K, Jiang, W, Zhang, Z.Y. | | 登録日 | 2020-09-16 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Cryo-electron microscopy structures of VCP/p97 reveal a new mechanism of oligomerization regulation.
Iscience, 24, 2021
|
|
7K56
 
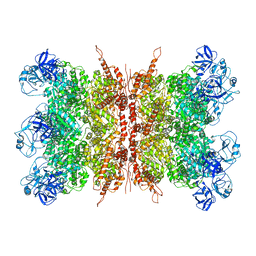 | | Structure of VCP dodecamer purified from H1299 cells | | 分子名称: | Transitional endoplasmic reticulum ATPase | | 著者 | Yu, G, Bai, Y, Li, K, Jiang, W, Zhang, Z.Y. | | 登録日 | 2020-09-16 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-electron microscopy structures of VCP/p97 reveal a new mechanism of oligomerization regulation.
Iscience, 24, 2021
|
|
7K57
 
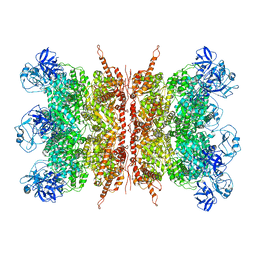 | | Structure of apo VCP dodecamer generated from bacterially recombinant VCP/p97 | | 分子名称: | Transitional endoplasmic reticulum ATPase | | 著者 | Yu, G, Bai, Y, Li, K, Jiang, W, Zhang, Z.Y. | | 登録日 | 2020-09-16 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-electron microscopy structures of VCP/p97 reveal a new mechanism of oligomerization regulation.
Iscience, 24, 2021
|
|
7ABR
 
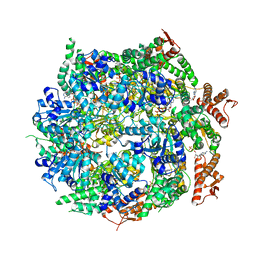 | | Cryo-EM structure of B. subtilis ClpC (DWB mutant) hexamer bound to a substrate polypeptide | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Negative regulator of genetic competence ClpC/MecB, ... | | 著者 | Morreale, F.E, Meinhart, A, Haselbach, D, Clausen, T. | | 登録日 | 2020-09-08 | | 公開日 | 2021-10-06 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
7D5T
 
 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state F1) | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | 著者 | Du, Y, Zhang, J, An, W, Ye, K. | | 登録日 | 2020-09-28 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | Cryo-EM structure of 90S preribosome with inactive Utp24 (state F1)
To Be Published
|
|
7D4I
 
 | | Cryo-EM structure of 90S small ribosomal precursors complex with the DEAH-box RNA helicase Dhr1 (State F) | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | 著者 | Du, Y, Zhang, J, An, W, Ye, K. | | 登録日 | 2020-09-24 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Cryo-EM structure of 90S small ribosomal precursors complex with Dhr1
To Be Published
|
|
7D63
 
 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state C) | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | 著者 | Du, Y, Zhang, J, An, W, Ye, K. | | 登録日 | 2020-09-29 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (12.3 Å) | | 主引用文献 | Cryo-EM structure of 90S preribosome with inactive Utp24 (state C)
To Be Published
|
|
7D5S
 
 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state A2) | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S12, ... | | 著者 | Du, Y, Zhang, J, An, W, Ye, K. | | 登録日 | 2020-09-28 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Cryo-EM structure of 90S preribosome with inactive Utp24 (state A2)
To Be Published
|
|
