3LE4
 
 | |
1AO4
 
 | | COBALT(III)-PEPLOMYCIN COMPLEX DETERMINED BY NMR STUDIES | | 分子名称: | 3-O-carbamoyl-alpha-D-mannopyranose-(1-2)-alpha-L-gulopyranose, AGLYCON OF PEPLOMYCIN, COBALT (III) ION, ... | | 著者 | Caceres-Cortes, J, Sugiyama, H, Ikudome, K, Saito, I, Wang, A.H.-J. | | 登録日 | 1997-07-16 | | 公開日 | 1999-07-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of cobalt(III)-pepleomycin and cobalt(III)-deglycopepleomycin (green forms) determined by NMR studies.
Eur.J.Biochem., 244, 1997
|
|
3LK4
 
 | | Crystal structure of CapZ bound to the uncapping motif from CD2AP | | 分子名称: | CD2-associated protein, F-actin-capping protein subunit alpha-1, F-actin-capping protein subunit beta isoforms 1 and 2 | | 著者 | Hernandez-Valladares, M, Kim, T, Kannan, B, Tung, A, Cooper, J.A, Robinson, R.C. | | 登録日 | 2010-01-27 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structural characterization of a capping protein interaction motif defines a family of actin filament regulators.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3LJW
 
 | | Crystal Structure of the Second Bromodomain of Human Polybromo | | 分子名称: | ACETATE ION, Protein polybromo-1, SODIUM ION | | 著者 | Charlop-Powers, Z, Zhou, M.M, Zeng, L, Zhang, Q. | | 登録日 | 2010-01-26 | | 公開日 | 2010-05-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.501 Å) | | 主引用文献 | Structural insights into selective histone H3 recognition by the human Polybromo bromodomain 2.
Cell Res., 20, 2010
|
|
3L6V
 
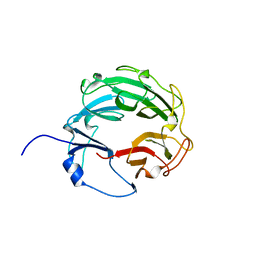 | | Crystal Structure of the Xanthomonas campestris Gyrase A C-terminal Domain | | 分子名称: | DNA gyrase subunit A | | 著者 | Hsieh, T.J, Yen, T.J, Lin, T.S, Chang, H.T, Huang, S.Y, Farh, L, Chan, N.L. | | 登録日 | 2009-12-26 | | 公開日 | 2010-03-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Twisting of the DNA binding surface by a beta-strand-bearing proline modulates DNA gyrase activity
To be Published
|
|
3L7P
 
 | |
6RV8
 
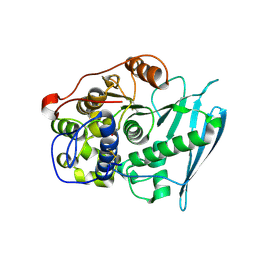 | | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor covalent complex with the aldouronic acid UXXR | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-Xylitol, ... | | 著者 | Ernst, H.A, Mosbech, C, Langkilde, A, Westh, P, Meyer, A, Agger, J.W, Larsen, S. | | 登録日 | 2019-05-31 | | 公開日 | 2020-03-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun, 11, 2020
|
|
3LP5
 
 | |
3LPB
 
 | | Crystal structure of Jak2 complexed with a potent 2,8-diaryl-quinoxaline inhibitor | | 分子名称: | N-methyl-4-[3-(3,4,5-trimethoxyphenyl)quinoxalin-5-yl]benzenesulfonamide, Tyrosine-protein kinase JAK2 | | 著者 | Tavares, G.A, Pissot-Soldermann, C, Gerspacher, M, Furet, P, Kroemer, M. | | 登録日 | 2010-02-05 | | 公開日 | 2010-04-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery and SAR of potent, orally available 2,8-diaryl-quinoxalines as a new class of JAK2 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LQH
 
 | |
3LAX
 
 | |
3LB0
 
 | | Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2 with Citrate Bound to the Active Site. | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-dehydroquinate dehydratase, CITRIC ACID, ... | | 著者 | Minasov, G, Light, S.H, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-01-07 | | 公開日 | 2010-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2 with Citrate Bound to the Active Site.
TO BE PUBLISHED
|
|
3LBK
 
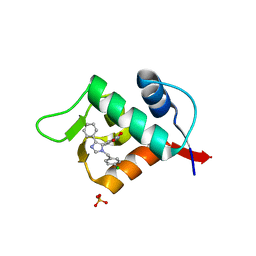 | | Structure of human MDM2 protein in complex with a small molecule inhibitor | | 分子名称: | 6-chloro-3-[1-(4-chlorobenzyl)-4-phenyl-1H-imidazol-5-yl]-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | 著者 | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | 登録日 | 2010-01-08 | | 公開日 | 2010-03-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
3LBO
 
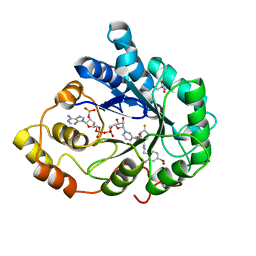 | | Human aldose reductase mutant T113C complexed with IDD594 | | 分子名称: | Aldose reductase, BROMIDE ION, CITRIC ACID, ... | | 著者 | Koch, C, Heine, A, Klebe, G. | | 登録日 | 2010-01-08 | | 公開日 | 2010-12-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Tracing the detail: how mutations affect binding modes and thermodynamic signatures of closely related aldose reductase inhibitors
J.Mol.Biol., 406, 2011
|
|
4WRN
 
 | | Crystal structure of the polymerization region of human uromodulin/Tamm-Horsfall protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltose-binding periplasmic protein,Uromodulin, ZINC ION, ... | | 著者 | Bokhove, M, De Sanctis, D, Jovine, L. | | 登録日 | 2014-10-24 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | A structured interdomain linker directs self-polymerization of human uromodulin.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6S0B
 
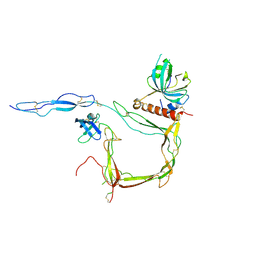 | | Crystal Structure of Properdin in complex with the CTC domain of C3/C3b | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3, Properdin, ... | | 著者 | van den Bos, R.M, Pearce, N.M, Gros, P. | | 登録日 | 2019-06-14 | | 公開日 | 2019-09-04 | | 最終更新日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (2.312 Å) | | 主引用文献 | Insights Into Enhanced Complement Activation by Structures of Properdin and Its Complex With the C-Terminal Domain of C3b.
Front Immunol, 10, 2019
|
|
6S0F
 
 | | Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with 3-Deoxy-D-glycero-D-galacto-2-nonulosonic acid | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | 著者 | Bule, P, Blagova, E, Chuzel, L, Taron, C.H, Davies, G.J. | | 登録日 | 2019-06-14 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Inverting family GH156 sialidases define an unusual catalytic motif for glycosidase action.
Nat Commun, 10, 2019
|
|
6LXJ
 
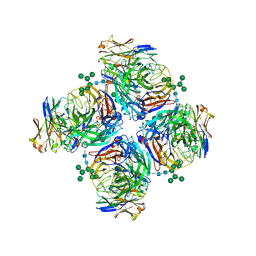 | | Crystal structure of human Z2B3 Fab in complex with influenza virus neuraminidase from A/Anhui/1/2013 (H7N9) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Heavy chain of Z2B3 Fab, ... | | 著者 | Jiang, H, Peng, W, Qi, J, Chai, Y, Song, H, Shi, Y, Gao, G.F, Wu, Y. | | 登録日 | 2020-02-11 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.903 Å) | | 主引用文献 | Structure-Based Modification of an Anti-neuraminidase Human Antibody Restores Protection Efficacy against the Drifted Influenza Virus.
Mbio, 11, 2020
|
|
1BFC
 
 | | BASIC FIBROBLAST GROWTH FACTOR COMPLEXED WITH HEPARIN HEXAMER FRAGMENT | | 分子名称: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, BASIC FIBROBLAST GROWTH FACTOR | | 著者 | Faham, S, Rees, D.C. | | 登録日 | 1995-12-12 | | 公開日 | 1996-04-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Heparin structure and interactions with basic fibroblast growth factor.
Science, 271, 1996
|
|
7Q1K
 
 | | Crystal structure of the native AA9A LPMO from Thermoascus aurantiacus | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, GLYCEROL, ... | | 著者 | Yu, W, Mohsin, I, Li, D.C, Papageorgiou, A.C. | | 登録日 | 2021-10-20 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Purification and Structural Characterization of the Auxiliary Activity 9 Native Lytic Polysaccharide Monooxygenase from Thermoascus aurantiacus and Identification of Its C1- and C4-Oxidized Reaction Products
Catalysts, 12, 2022
|
|
1BCX
 
 | |
5KXD
 
 | | Wisteria floribunda lectin in complex with GalNAc(beta1-4)GlcNAc (LacdiNAc) at pH 6.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | 著者 | Evans, S.V, Haji-Ghassemi, O. | | 登録日 | 2016-07-20 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Molecular Basis for Recognition of the Cancer Glycobiomarker, LacdiNAc (GalNAc[ beta 14]GlcNAc), by Wisteria floribunda Agglutinin.
J.Biol.Chem., 291, 2016
|
|
7N18
 
 | | Clostridium botulinum Neurotoxin Serotype A Light Chain Inhibited by a Chiral Hydroxamic Acid | | 分子名称: | (3R)-3-(4-chlorophenyl)-N,5-dihydroxypentanamide, (3S)-3-(4-chlorophenyl)-N,5-dihydroxypentanamide, Botulinum neurotoxin type A, ... | | 著者 | Silvaggi, N.R, Allen, K.N. | | 登録日 | 2021-05-27 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Use of Crystallography and Molecular Modeling for the Inhibition of the Botulinum Neurotoxin A Protease.
Acs Med.Chem.Lett., 12, 2021
|
|
1B9Z
 
 | | BACILLUS CEREUS BETA-AMYLASE COMPLEXED WITH MALTOSE | | 分子名称: | ACETATE ION, CALCIUM ION, PROTEIN (BETA-AMYLASE), ... | | 著者 | Mikami, B, Adachi, M, Kage, T, Sarikaya, E, Nanmori, T, Shinke, R, Utsumi, S. | | 登録日 | 1999-03-06 | | 公開日 | 1999-03-15 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of raw starch-digesting Bacillus cereus beta-amylase complexed with maltose.
Biochemistry, 38, 1999
|
|
6SEA
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q in complex with lactose bound in deep mode | | 分子名称: | ACETATE ION, Beta-galactosidase, SODIUM ION, ... | | 著者 | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | 登録日 | 2019-07-29 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.869 Å) | | 主引用文献 | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
