1KDL
 
 | |
4E2T
 
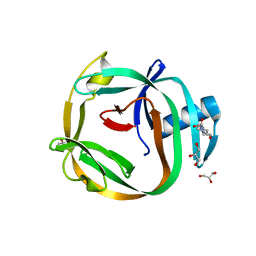 | | Crystal Structures of RadA intein from Pyrococcus horikoshii | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Pho radA intein | | 著者 | Oeemig, J.S, Zhou, D, Kajander, T, Wlodawer, A, Iwai, H. | | 登録日 | 2012-03-09 | | 公開日 | 2012-05-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | NMR and Crystal Structures of the Pyrococcus horikoshii RadA Intein Guide a Strategy for Engineering a Highly Efficient and Promiscuous Intein.
J.Mol.Biol., 421, 2012
|
|
4E2U
 
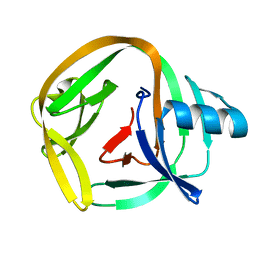 | | Crystal Structures of RadAmin intein from Pyrococcus horikoshii | | 分子名称: | Pho radA intein | | 著者 | Oeemig, J.S, Zhou, D, Kajander, T, Wlodawer, A, Iwai, H. | | 登録日 | 2012-03-09 | | 公開日 | 2012-05-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.582 Å) | | 主引用文献 | NMR and Crystal Structures of the Pyrococcus horikoshii RadA Intein Guide a Strategy for Engineering a Highly Efficient and Promiscuous Intein.
J.Mol.Biol., 421, 2012
|
|
1IHQ
 
 | | GLYTM1BZIP: A CHIMERIC PEPTIDE MODEL OF THE N-TERMINUS OF A RAT SHORT ALPHA TROPOMYOSIN WITH THE N-TERMINUS ENCODED BY EXON 1B | | 分子名称: | CHIMERIC PEPTIDE GlyTM1bZip: TROPOMYOSIN ALPHA CHAIN, BRAIN-3 and GENERAL CONTROL PROTEIN GCN4 | | 著者 | Greenfield, N.J, Yuang, Y.J, Palm, T, Swapna, G.V, Monleon, D, Montelione, G.T, Hitchcock-Degregori, S.E, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2001-04-19 | | 公開日 | 2001-10-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure and folding dynamics of the N terminus of a rat non-muscle alpha-tropomyosin in an engineered chimeric protein.
J.Mol.Biol., 312, 2001
|
|
1NOQ
 
 | | e-motif structure | | 分子名称: | 5'-D(*CP*CP*GP*CP*CP*G)-3' | | 著者 | Zheng, M, Huang, X, Smith, G.K, Yang, X, Gao, X. | | 登録日 | 2003-01-16 | | 公開日 | 2003-02-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Genetically unstable CXG repeats are structurally dynamic and have a high propensity for folding.
An NMR and UV spectroscopic study.
J.Mol.Biol., 264, 1996
|
|
1L5E
 
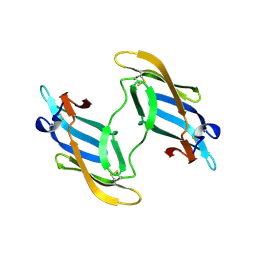 | | The domain-swapped dimer of CV-N in solution | | 分子名称: | Cyanovirin-N | | 著者 | Barrientos, L.G, Louis, J.M, Botos, I, Mori, T, Han, Z, O'Keefe, B.R, Boyd, M.R, Wlodawer, A, Gronenborn, A.M. | | 登録日 | 2002-03-06 | | 公開日 | 2002-06-05 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The domain-swapped dimer of cyanovirin-N is in a metastable folded state: reconciliation of X-ray and NMR structures.
Structure, 10, 2002
|
|
2JQ0
 
 | | Phylloseptin-1 | | 分子名称: | Phylloseptin-1 | | 著者 | Resende, J.M, Mendonca Moraes, C, Almeida, F.C.L, Prates, M.V, Cesar, A, Valente, A, Bemquerer, M.P, Pilo-Veloso, D, Bechinger, B. | | 登録日 | 2007-05-25 | | 公開日 | 2008-06-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structures of the antimicrobial peptides phylloseptin-1, -2, and -3 and biological activity: The role of charges and hydrogen bonding interactions in stabilizing helix conformations
Peptides, 29, 2008
|
|
2KCZ
 
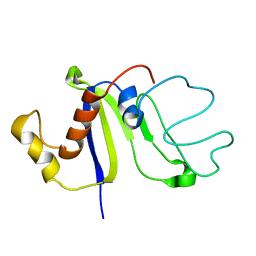 | | Solution NMR structure of the C-terminal domain of protein DR_A0006 from Deinococcus radiodurans. Northeast Structural Genomics Consortium Target DrR147D | | 分子名称: | Uncharacterized protein DR_A0006 | | 著者 | Mills, J.L, Ghosh, A, Garcia, E, Wang, H, Ciccosanti, C, Xiao, R, Nair, R, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-12-31 | | 公開日 | 2009-01-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure of the C-Terminal Domain of Protein DR_A0006 from Deinococcus radiodurans. Northeast Structural Genomics Consortium Target DrR147D.
To be Published
|
|
2KM0
 
 | | Cu(I)-bound CopK | | 分子名称: | COPPER (I) ION, Copper resistance protein K | | 著者 | Bersch, B. | | 登録日 | 2009-07-15 | | 公開日 | 2010-03-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | CopK from Cupriavidus metallidurans CH34 Binds Cu(I) in a Tetrathioether Site: Characterization by X-ray Absorption and NMR Spectroscopy
J.Am.Chem.Soc., 2010
|
|
2KEY
 
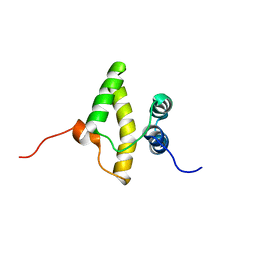 | | Solution NMR structure of a domain from a putative phage integrase protein BF2284 from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR257C | | 分子名称: | Putative phage integrase | | 著者 | Mills, J.L, Sathyamoorthy, B, Sukumaran, D.K, Lee, D, Ciccosanti, C, Jiang, M, Xiao, R, Acton, T.B, Swapna, G.V.T, Rost, B, Nair, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2009-02-06 | | 公開日 | 2009-04-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure of a Domain from a Putative Phage Integrase Protein, BF2284, from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR257C
To be Published
|
|
1DK9
 
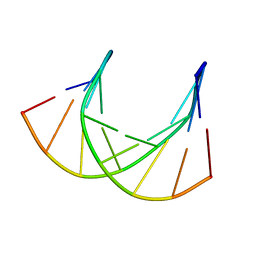 | | SOLUTION STRUCTURE ANALYSIS OF THE DNA DUPLEX D(CATGAGTAC)D(GTACTCATG) | | 分子名称: | 5'-D(CP*AP*TP*GP*AP*GP*TP*AP*CP*)-3', 5'-D(GP*TP*AP*CP*TP*CP*AP*TP*GP*)-3' | | 著者 | Klewer, D.A, Hoskins, A, Davisson, V.J, Bergstrom, D.E, LiWang, A.C. | | 登録日 | 1999-12-06 | | 公開日 | 1999-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of a DNA duplex containing nucleoside analog 1-(2'-deoxy-beta-D-ribofuranosyl)-3-nitropyrrole and the structure of the unmodified control.
Nucleic Acids Res., 28, 2000
|
|
1CI5
 
 | | GLYCAN-FREE MUTANT ADHESION DOMAIN OF HUMAN CD58 (LFA-3) | | 分子名称: | PROTEIN (LYMPHOCYTE FUNCTION-ASSOCIATED ANTIGEN 3(CD58)) | | 著者 | Sun, Z.Y.J, Dotsch, V, Kim, M, Li, J, Reinherz, E.L, Wagner, G. | | 登録日 | 1999-04-07 | | 公開日 | 1999-06-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Functional glycan-free adhesion domain of human cell surface receptor CD58: design, production and NMR studies.
EMBO J., 18, 1999
|
|
5MVB
 
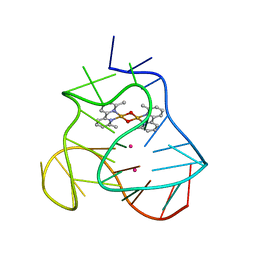 | | Solution structure of a human G-Quadruplex hybrid-2 form in complex with a Gold-ligand. | | 分子名称: | DNA (26-MER), POTASSIUM ION, bis(m2-Oxo)-bis(2-methyl-2,2'-bipyridine)-di-gold(iii) | | 著者 | Wirmer-Bartoschek, J, Jonker, H.R.A, Bendel, L.E, Gruen, T, Bazzicalupi, C, Messori, L, Gratteri, P, Schwalbe, H. | | 登録日 | 2017-01-16 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure of a Ligand/Hybrid-2-G-Quadruplex Complex Reveals Rearrangements that Affect Ligand Binding.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
1ATY
 
 | |
6ROA
 
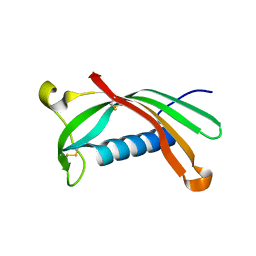 | | Crystal structure of V57G mutant of human cystatin C | | 分子名称: | Cystatin-C | | 著者 | Orlikowska, M, Behrendt, I, Borek, D, Otwinowski, Z, Skowron, P, Szymanska, A. | | 登録日 | 2019-05-10 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | NMR and crystallographic structural studies of the extremely stable monomeric variant of human cystatin C with single amino acid substitution.
Febs J., 287, 2020
|
|
2JQN
 
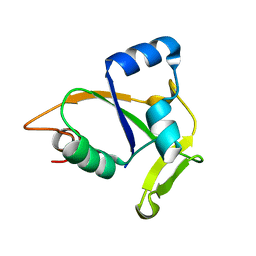 | | Solution NMR structure of CC0527 from Caulobacter crescentus. Northeast Structural Genomics Target CCR55 | | 分子名称: | Hypothetical protein | | 著者 | Aramini, J.M, Rossi, P, Moseley, H.N.B, Wang, D, Nwosu, C, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-06-05 | | 公開日 | 2007-07-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of CC0527 from Caulobacter crescentus
To be Published
|
|
6IYN
 
 | | Solution structure of camelid nanobody Nb26 against aflatoxin B1 | | 分子名称: | Nb26 | | 著者 | Nie, Y, He, T, Zhu, J, Li, S.L, Hu, R, Yang, Y.H. | | 登録日 | 2018-12-17 | | 公開日 | 2019-01-23 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Chemical shift assignments of a camelid nanobody against aflatoxin B1.
Biomol NMR Assign, 13, 2019
|
|
1R05
 
 | | Solution Structure of Max B-HLH-LZ | | 分子名称: | Max protein | | 著者 | Sauv, S, Tremblay, L, Lavigne, P. | | 登録日 | 2003-09-19 | | 公開日 | 2003-10-21 | | 最終更新日 | 2021-10-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The NMR solution structure of a mutant of the Max b/HLH/LZ free of DNA: insights into the specific and reversible DNA binding mechanism of dimeric transcription factors
J.Mol.Biol., 342, 2004
|
|
6I1W
 
 | | Structure of the RNA duplex containing pseudouridine residue (5'-Gp(PSU)pC-3' sequence context) | | 分子名称: | RNA (5'-R(*AP*CP*UP*GP*AP*CP*UP*GP*A)-3'), RNA (5'-R(*UP*CP*AP*GP*(PSU)P*CP*AP*GP*U)-3') | | 著者 | Deb, I, Popenda, L, Sarzynska, J, Gdaniec, Z. | | 登録日 | 2018-10-30 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Computational and NMR studies of RNA duplexes with an internal pseudouridine-adenosine base pair.
Sci Rep, 9, 2019
|
|
6I1V
 
 | | Structure of the RNA duplex containing pseudouridine residue (5'-Cp(PSU)pG-3' sequence context) | | 分子名称: | RNA (5'-R(*AP*CP*UP*CP*AP*GP*UP*GP*A)-3'), RNA (5'-R(*UP*CP*AP*CP*(PSU)P*GP*AP*GP*U)-3') | | 著者 | Deb, I, Popenda, L, Sarzynska, J, Gdaniec, Z. | | 登録日 | 2018-10-30 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Computational and NMR studies of RNA duplexes with an internal pseudouridine-adenosine base pair.
Sci Rep, 9, 2019
|
|
1HJ0
 
 | | Thymosin beta9 | | 分子名称: | THYMOSIN BETA9 | | 著者 | Stoll, R, Voelter, W, Holak, T.A. | | 登録日 | 2001-01-05 | | 公開日 | 2002-01-04 | | 最終更新日 | 2024-04-24 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Conformation of Thymosin Beta9 in Water/Fluoroalcohol Solution Determined by NMR Spectroscopy
Biopolymers, 41, 1997
|
|
5TTT
 
 | | Sparse-restraint solution NMR structure of micelle-solubilized cytosolic amino terminal domain of C. elegans mechanosensory ion channel MEC-4 refined by restrained Rosetta | | 分子名称: | Degenerin mec-4 | | 著者 | Everett, J.K, Liu, G, Mao, B, Driscoll, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2016-11-04 | | 公開日 | 2017-02-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Sparse-restraint solution NMR structure of micelle-solubilized
cytosolic amino terminal domain of C. elegans mechanosensory ion channel
MEC-4 refined by restrained Rosetta
To Be Published
|
|
1LSI
 
 | |
1JCO
 
 | | Solution structure of the monomeric [Thr(B27)->Pro,Pro(B28)->Thr] insulin mutant (PT insulin) | | 分子名称: | Insulin A chain, Insulin B chain | | 著者 | Keller, D, Clausen, R, Josefsen, K, Led, J.J. | | 登録日 | 2001-06-11 | | 公開日 | 2001-10-03 | | 最終更新日 | 2021-10-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Flexibility and bioactivity of insulin: an NMR investigation of the solution structure and folding of an unusually flexible human insulin mutant with increased biological activity.
Biochemistry, 40, 2001
|
|
1OJG
 
 | | Sensory domain of the membraneous two-component fumarate sensor DcuS of E. coli | | 分子名称: | SENSOR PROTEIN DCUS | | 著者 | Pappalardo, L, Janausch, I.G, Vijayan, V, Zientz, E, Junker, J, Peti, W, Zweckstetter, M, Unden, G, Griesinger, C. | | 登録日 | 2003-07-10 | | 公開日 | 2003-08-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The NMR structure of the sensory domain of the membranous two-component fumarate sensor (histidine protein kinase) DcuS of Escherichia coli.
J. Biol. Chem., 278, 2003
|
|
