5FNR
 
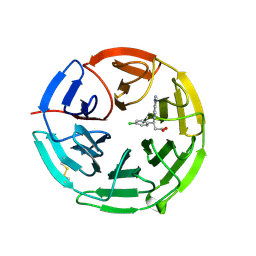 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | 分子名称: | (3S)-3-(4-chlorophenyl)-3-(1-methylbenzotriazol-5-yl)propanoic acid, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | 著者 | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | 登録日 | 2015-11-16 | | 公開日 | 2016-04-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FNT
 
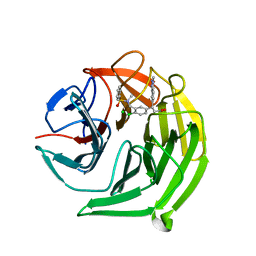 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | 分子名称: | (3S)-3-{4-Chloro-3-[(N-methylbenzenesulfonamido) methyl]phenyl}-3-(1-methyl-1H-1,2,3-benzotriazol-5-yl)propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | 著者 | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | 登録日 | 2015-11-16 | | 公開日 | 2016-04-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FZJ
 
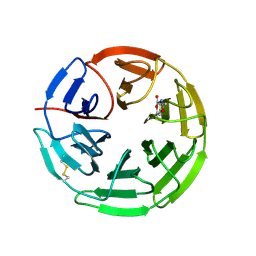 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | 分子名称: | 2,6-DIMETHYL-4H-PYRANO[3,4-D][1,3]OXAZOL-4-ONE, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | 著者 | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | 登録日 | 2016-03-14 | | 公開日 | 2016-04-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
6F6Y
 
 | |
1DKY
 
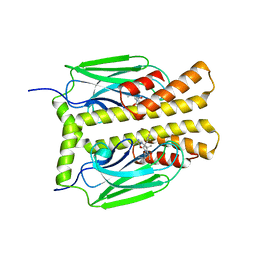 | | THE SUBSTRATE BINDING DOMAIN OF DNAK IN COMPLEX WITH A SUBSTRATE PEPTIDE, DETERMINED FROM TYPE 2 NATIVE CRYSTALS | | 分子名称: | DNAK, PEPTIDE SUBSTRATE | | 著者 | Zhu, X, Zhao, X, Burkholder, W.F, Gragerov, A, Ogata, C.M, Gottesman, M.E, Hendrickson, W.A. | | 登録日 | 1996-06-03 | | 公開日 | 1996-12-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural analysis of substrate binding by the molecular chaperone DnaK.
Science, 272, 1996
|
|
1DKX
 
 | | THE SUBSTRATE BINDING DOMAIN OF DNAK IN COMPLEX WITH A SUBSTRATE PEPTIDE, DETERMINED FROM TYPE 1 SELENOMETHIONYL CRYSTALS | | 分子名称: | SUBSTRATE BINDING DOMAIN OF DNAK, SUBSTRATE PEPTIDE (7 RESIDUES) | | 著者 | Zhu, X, Zhao, X, Burkholder, W.F, Gragerov, A, Ogata, C.M, Gottesman, M.E, Hendrickson, W.A. | | 登録日 | 1996-06-03 | | 公開日 | 1996-12-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural analysis of substrate binding by the molecular chaperone DnaK.
Science, 272, 1996
|
|
5FNQ
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | 分子名称: | 3-(4-CHLOROPHENYL)PROPANOIC ACID, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | 著者 | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | 登録日 | 2015-11-16 | | 公開日 | 2016-04-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
1D4F
 
 | | CRYSTAL STRUCTURE OF RECOMBINANT RAT-LIVER D244E MUTANT S-ADENOSYLHOMOCYSTEINE HYDROLASE | | 分子名称: | ADENOSINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-ADENOSYLHOMOCYSTEINE HYDROLASE | | 著者 | Komoto, J, Huang, Y, Takusagawa, F, Gomi, T, Ogawa, H, Takata, Y, Fujioka, M. | | 登録日 | 2000-06-22 | | 公開日 | 2001-01-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Effects of site-directed mutagenesis on structure and function of recombinant rat liver S-adenosylhomocysteine hydrolase. Crystal structure of D244E mutant enzyme.
J.Biol.Chem., 275, 2000
|
|
7UM3
 
 | | Crystal structure of a Fab in complex with a peptide derived from the LAG-3 D1 domain loop insertion | | 分子名称: | D1 domain loop peptide from Lymphocyte activation gene 3 protein, Fab heavy chain, Fab light chain | | 著者 | Zorn, J.A, Lee, P.S, Rajpal, A, Strop, P. | | 登録日 | 2022-04-06 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3983 Å) | | 主引用文献 | Preclinical Characterization of Relatlimab, a Human LAG-3-Blocking Antibody, Alone or in Combination with Nivolumab.
Cancer Immunol Res, 10, 2022
|
|
7VMY
 
 | | Crystal structure of LimF prenyltransferase bound with GSPP | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GERANYL S-THIOLODIPHOSPHATE, LynF/TruF/PatF family peptide O-prenyltransferase, ... | | 著者 | Hamada, K, Kobayashi, S, Okada, C, Zhang, Y, Inoue, S, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | 登録日 | 2021-10-09 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | LimF is a versatile prenyltransferase for histidine-C-geranylation on diverse non-natural substrates
Nat Catal, 2022
|
|
7VMW
 
 | | Crystal structure of LimF prenyltransferase bound with a peptide substrate and GSPP | | 分子名称: | GERANYL S-THIOLODIPHOSPHATE, LynF/TruF/PatF family peptide O-prenyltransferase, MAGNESIUM ION, ... | | 著者 | Hamada, K, Kobayashi, S, Okada, C, Zhang, Y, Inoue, S, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | 登録日 | 2021-10-09 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | LimF is a versatile prenyltransferase for histidine-C-geranylation on diverse non-natural substrates
Nat Catal, 2022
|
|
6G0C
 
 | | Crystal structure of SdeA catalytic core | | 分子名称: | 1,2-ETHANEDIOL, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, Ubiquitinating/deubiquitinating enzyme SdeA | | 著者 | Kalayil, S, Bhogaraju, S, Basquin, J, Dikic, I. | | 登録日 | 2018-03-17 | | 公開日 | 2018-05-30 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.802 Å) | | 主引用文献 | Insights into catalysis and function of phosphoribosyl-linked serine ubiquitination.
Nature, 557, 2018
|
|
6FM2
 
 | | CARP domain of mouse cyclase-associated protein 1 (CAP1) bound to ADP-actin | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Kotila, T.M, Kogan, K, Lappalainen, P. | | 登録日 | 2018-01-30 | | 公開日 | 2018-05-16 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis of actin monomer re-charging by cyclase-associated protein.
Nat Commun, 9, 2018
|
|
1GIQ
 
 | | Crystal Structure of the Enzymatic Componet of Iota-Toxin from Clostridium Perfringens with NADH | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, IOTA TOXIN COMPONENT IA | | 著者 | Tsuge, H, Nagahama, M, Nishimura, H, Hisatsune, J, Sakaguchi, Y, Itogawa, Y, Katunuma, N, Sakurai, J. | | 登録日 | 2001-03-12 | | 公開日 | 2003-01-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure and Site-directed Mutagenesis of Enzymatic Components from Clostridium perfringens Iota-toxin
J.MOL.BIOL., 325, 2003
|
|
6H8K
 
 | | Crystal structure of a variant (Q133C in PSST) of Yarrowia lipolytica complex I | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, NADH dehydrogenase [ubiquinone] flavoprotein 1, ... | | 著者 | Wirth, C, Galemou Yoga, E, Zickermann, V, Hunte, C. | | 登録日 | 2018-08-02 | | 公開日 | 2018-12-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.79 Å) | | 主引用文献 | Locking loop movement in the ubiquinone pocket of complex I disengages the proton pumps.
Nat Commun, 9, 2018
|
|
4LZF
 
 | | A novel domain in the microcephaly protein CPAP suggests a role in centriole architecture | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Centrosomal P4.1-associated protein, SCL-interrupting locus protein homolog, ... | | 著者 | Hatzopoulos, G.N, Erat, M.C, Cutts, E, Rogala, K, Slatter, L, Stansfeld, P.J, Vakonakis, I. | | 登録日 | 2013-07-31 | | 公開日 | 2013-09-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Structural analysis of the G-box domain of the microcephaly protein CPAP suggests a role in centriole architecture.
Structure, 21, 2013
|
|
6G71
 
 | | Structure of CYP1232A24 from Arthrobacter sp. | | 分子名称: | 1,2-ETHANEDIOL, Cytochrome P450, FE (III) ION, ... | | 著者 | Dubiel, P, Sharma, M, Klenk, J, Hauer, B, Grogan, G. | | 登録日 | 2018-04-04 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Identification and characterization of cytochrome P450 1232A24 and 1232F1 from Arthrobacter sp. and their role in the metabolic pathway of papaverine.
J.Biochem., 166, 2019
|
|
7X5F
 
 | | Nrf2-MafG heterodimer bound with CsMBE2 | | 分子名称: | Nuclear factor erythroid 2-related factor 2, Synthetic DNA, Transcription factor MafG | | 著者 | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | 登録日 | 2022-03-04 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
7X5G
 
 | | Nrf2 (A510Y)-MafG heterodimer bound with CsMBE2 | | 分子名称: | DNA (5'-D(*CP*AP*CP*AP*GP*TP*GP*AP*CP*TP*CP*AP*GP*CP*AP*G)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*TP*GP*AP*GP*TP*CP*AP*CP*TP*GP*T)-3'), Nuclear factor erythroid 2-related factor 2, ... | | 著者 | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | 登録日 | 2022-03-04 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
7X5E
 
 | | Nrf2-MafG heterodimer bound with CsMBE1 | | 分子名称: | DNA (5'-D(*CP*AP*TP*GP*AP*TP*GP*AP*GP*TP*CP*AP*GP*CP*AP*A)-3'), DNA (5'-D(*GP*TP*TP*GP*CP*TP*GP*AP*CP*TP*CP*AP*TP*CP*AP*T)-3'), HEXAETHYLENE GLYCOL, ... | | 著者 | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | 登録日 | 2022-03-04 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
1GC5
 
 | | CRYSTAL STRUCTURE OF A NOVEL ADP-DEPENDENT GLUCOKINASE FROM THERMOCOCCUS LITORALIS | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADP-DEPENDENT GLUCOKINASE | | 著者 | Ito, S, Fushinobu, S, Yoshioka, I, Koga, S, Matsuzawa, H, Wakagi, T. | | 登録日 | 2000-07-20 | | 公開日 | 2001-07-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Basis for the ADP-Specificity of a Novel Glucokinase from a Hyperthermophilic Archaeon
Structure, 9, 2001
|
|
1GIR
 
 | | CRYSTAL STRUCTURE OF THE ENZYMATIC COMPONET OF IOTA-TOXIN FROM CLOSTRIDIUM PERFRINGENS WITH NADPH | | 分子名称: | IOTA TOXIN COMPONENT IA, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Tsuge, H, Nagahama, M, Nishimura, H, Hisatsune, J, Sakaguchi, Y, Itogawa, Y, Katunuma, N, Sakurai, J. | | 登録日 | 2001-03-12 | | 公開日 | 2003-01-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure and Site-directed Mutagenesis of Enzymatic Components from Clostridium perfringens Iota-toxin
J.MOL.BIOL., 325, 2003
|
|
3O6R
 
 | | Crystal Structure of 4-Chlorocatechol Dioxygenase from Rhodococcus opacus 1CP in complex with pyrogallol | | 分子名称: | (2R)-3-(PHOSPHONOOXY)-2-(TETRADECANOYLOXY)PROPYL PALMITATE, BENZENE-1,2,3-TRIOL, Chlorocatechol 1,2-dioxygenase, ... | | 著者 | Ferraroni, M, Briganti, F, Kolomitseva, M, Golovleva, L. | | 登録日 | 2010-07-29 | | 公開日 | 2011-08-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | X-ray structures of 4-chlorocatechol 1,2-dioxygenase adducts with substituted catechols: new perspectives in the molecular basis of intradiol ring cleaving dioxygenases specificity.
J. Struct. Biol., 181, 2013
|
|
6GIO
 
 | |
1H2R
 
 | |
