1IAG
 
 | |
4BI1
 
 | | Scaffold Focused Virtual Screening: Prospective Application to the Discovery of TTK Inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, DUAL SPECIFICITY PROTEIN KINASE TTK, ... | | 著者 | Langdon, S.R, Westwood, I.M, van Montfort, R.L.M, Brown, N, Blagg, J. | | 登録日 | 2013-04-09 | | 公開日 | 2013-05-22 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Scaffold-Focused Virtual Screening: Prospective Application to the Discovery of Ttk Inhibitors.
J.Chem.Inf.Model, 53, 2013
|
|
2G6Q
 
 | |
3LE4
 
 | |
2JHB
 
 | |
2F6X
 
 | |
1B5F
 
 | | NATIVE CARDOSIN A FROM CYNARA CARDUNCULUS L. | | 分子名称: | PROTEIN (CARDOSIN A), alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Frazao, C, Bento, I, Carrondo, M.A. | | 登録日 | 1999-01-06 | | 公開日 | 1999-01-13 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Crystal structure of cardosin A, a glycosylated and Arg-Gly-Asp-containing aspartic proteinase from the flowers of Cynara cardunculus L.
J.Biol.Chem., 274, 1999
|
|
1DY4
 
 | | CBH1 IN COMPLEX WITH S-PROPRANOLOL | | 分子名称: | 1-(ISOPROPYLAMINO)-3-(1-NAPHTHYLOXY)-2-PROPANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | 著者 | Stahlberg, J, Henriksson, H, Divne, C, Isaksson, R, Pettersson, G, Johansson, G, Jones, T.A. | | 登録日 | 2000-01-26 | | 公開日 | 2000-12-18 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Basis for Enantiomer Binding and Separation of a Common Beta-Blocker: Crystal Structure of Cellobiohydrolase Cel7A with Bound (S)-Propranolol at 1.9 A Resolution
J.Mol.Biol., 305, 2001
|
|
4CZ1
 
 | | Crystal structure of kynurenine formamidase from Bacillus anthracis complexed with 2-aminoacetophenone. | | 分子名称: | 2-aminoacetophenone, KYNURENINE FORMAMIDASE, MAGNESIUM ION, ... | | 著者 | Diaz-Saez, L, Srikannathasan, V, Zoltner, M, Hunter, W.N. | | 登録日 | 2014-04-16 | | 公開日 | 2014-04-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structure of Bacterial Kynurenine Formamidase Reveals a Crowded Binuclear-Zinc Catalytic Site Primed to Generate a Potent Nucleophile.
Biochem.J., 462, 2014
|
|
1MPP
 
 | | X-RAY ANALYSES OF ASPARTIC PROTEINASES. V. STRUCTURE AND REFINEMENT AT 2.0 ANGSTROMS RESOLUTION OF THE ASPARTIC PROTEINASE FROM MUCOR PUSILLUS | | 分子名称: | PEPSIN, SULFATE ION | | 著者 | Newman, M, Watson, F, Roychowdhury, P, Jones, H, Badasso, M, Cleasby, A, Wood, S.P, Tickle, I.J, Blundell, T.L. | | 登録日 | 1992-02-19 | | 公開日 | 1993-10-31 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | X-ray analyses of aspartic proteinases. V. Structure and refinement at 2.0 A resolution of the aspartic proteinase from Mucor pusillus.
J.Mol.Biol., 230, 1993
|
|
2CKL
 
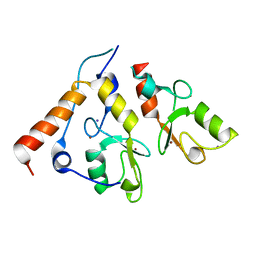 | | Ring1b-Bmi1 E3 catalytic domain structure | | 分子名称: | IODIDE ION, POLYCOMB GROUP RING FINGER PROTEIN 4, UBIQUITIN LIGASE PROTEIN RING2, ... | | 著者 | Buchwald, G, van der Stoop, P, Weichenrieder, O, Perrakis, A, van Lohuizen, M, Sixma, T.K. | | 登録日 | 2006-04-20 | | 公開日 | 2006-05-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure and E3-Ligase Activity of the Ring-Ring Complex of Polycomb Proteins Bmi1 and Ring1B.
Embo J., 25, 2006
|
|
2QX0
 
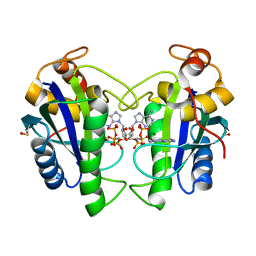 | | Crystal Structure of Yersinia pestis HPPK (Ternary Complex) | | 分子名称: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 7,8-dihydro-6-hydroxymethylpterin-pyrophosphokinase, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | 著者 | Blaszczyk, J, Cherry, S, Tropea, J.E, Waugh, D.S, Ji, X. | | 登録日 | 2007-08-10 | | 公開日 | 2007-10-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure and activity of Yersinia pestis 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase as a novel target for the development of antiplague therapeutics.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1XDF
 
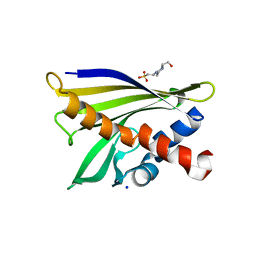 | | Crystal structure of pathogenesis-related protein LlPR-10.2A from yellow lupine | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PR10.2A, SODIUM ION | | 著者 | Pasternak, O, Biesiadka, J, Dolot, R, Handschuh, L, Bujacz, G, Sikorski, M.M, Jaskolski, M. | | 登録日 | 2004-09-06 | | 公開日 | 2005-02-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of a yellow lupin pathogenesis-related PR-10 protein belonging to a novel subclass.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1IFH
 
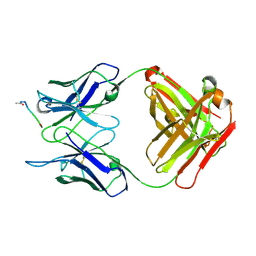 | |
6MSF
 
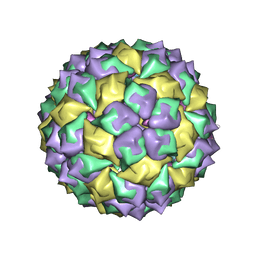 | | F6 APTAMER MS2 COAT PROTEIN COMPLEX | | 分子名称: | PROTEIN (MS2 PROTEIN CAPSID), RNA (5'-R(*CP*AP*GP*UP*CP*AP*CP*UP*GP*G)-3'), RNA (5'-R(*CP*CP*AP*CP*AP*GP*UP*CP*AP*CP*UP*GP*GP*G)-3') | | 著者 | Convery, M.A, Rowsell, S, Stonehouse, N.J, Ellington, A.D, Hirao, I, Murray, J.B, Peabody, D.S, Phillips, S.E.V, Stockley, P.G. | | 登録日 | 1998-01-06 | | 公開日 | 1998-07-08 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of an RNA aptamer-protein complex at 2.8 A resolution.
Nat.Struct.Biol., 5, 1998
|
|
1BCI
 
 | | C2 DOMAIN OF CYTOSOLIC PHOSPHOLIPASE A2, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | CALCIUM ION, CYTOSOLIC PHOSPHOLIPASE A2 | | 著者 | Xu, G.Y, Mcdonagh, T, Yu, H.A, Nalefski, E.A, Clark, J.D, Cumming, D.A. | | 登録日 | 1998-04-30 | | 公開日 | 1998-11-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and membrane interactions of the C2 domain of cytosolic phospholipase A2.
J.Mol.Biol., 280, 1998
|
|
4BI2
 
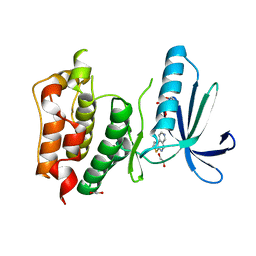 | | Scaffold Focused Virtual Screening: Prospective Application to the Discovery of TTK Inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, DUAL SPECIFICITY PROTEIN KINASE TTK, ... | | 著者 | Langdon, S.R, Westwood, I.M, van Montfort, R.L.M, Brown, N, Blagg, J. | | 登録日 | 2013-04-09 | | 公開日 | 2013-05-22 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | Scaffold-Focused Virtual Screening: Prospective Application to the Discovery of Ttk Inhibitors.
J.Chem.Inf.Model, 53, 2013
|
|
3GN1
 
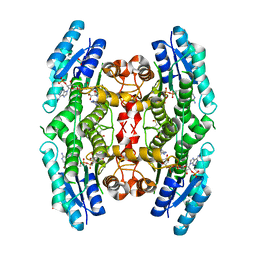 | | Structure of Pteridine Reductase 1 (PTR1) from TRYPANOSOMA BRUCEI in ternary complex with cofactor (NADP+) and inhibitor (DDD00067116) | | 分子名称: | 1H-benzimidazol-2-amine, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Tulloch, L.B, Brenk, R, Hunter, W.N. | | 登録日 | 2009-03-16 | | 公開日 | 2009-12-29 | | 最終更新日 | 2011-09-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | One scaffold, three binding modes: novel and selective pteridine reductase 1 inhibitors derived from fragment hits discovered by virtual screening.
J.Med.Chem., 52, 2009
|
|
4KGC
 
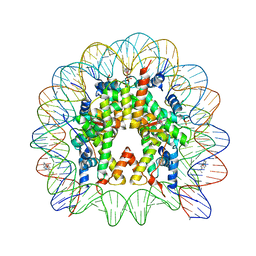 | | Nucleosome Core Particle Containing (ETA6-P-CYMENE)-(1, 2-ETHYLENEDIAMINE)-RUTHENIUM | | 分子名称: | (ethane-1,2-diamine-kappa~2~N,N')[(1,2,3,4,5,6-eta)-1-methyl-4-(propan-2-yl)cyclohexane-1,2,3,4,5,6-hexayl]ruthenium, DNA (145-mer), Histone H2A, ... | | 著者 | Adhireksan, Z, Davey, C.A. | | 登録日 | 2013-04-29 | | 公開日 | 2014-03-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Ligand substitutions between ruthenium-cymene compounds can control protein versus DNA targeting and anticancer activity
Nat Commun, 5, 2014
|
|
1N6Z
 
 | |
2BBV
 
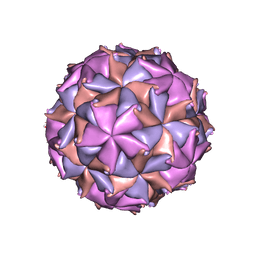 | | THE REFINED THREE-DIMENSIONAL STRUCTURE OF AN INSECT VIRUS AT 2.8 ANGSTROMS RESOLUTION | | 分子名称: | CALCIUM ION, PROTEIN (BLACK BEETLE VIRUS CAPSID PROTEIN), RNA (5'-R(*UP*CP*UP*UP*AP*UP*AP*UP*CP*U)-3') | | 著者 | Wery, J.-P, Reddy, V.S, Hosur, M.V, Johnson, J.E. | | 登録日 | 1994-06-06 | | 公開日 | 1994-08-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The refined three-dimensional structure of an insect virus at 2.8 A resolution.
J.Mol.Biol., 235, 1994
|
|
1E2Y
 
 | |
1DUD
 
 | |
1AH7
 
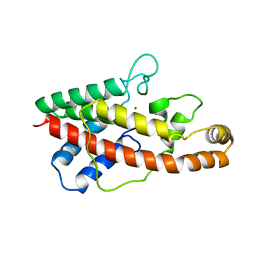 | | PHOSPHOLIPASE C FROM BACILLUS CEREUS | | 分子名称: | PHOSPHOLIPASE C, ZINC ION | | 著者 | Greaves, R. | | 登録日 | 1997-04-14 | | 公開日 | 1997-12-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.501 Å) | | 主引用文献 | High-resolution (1.5 A) crystal structure of phospholipase C from Bacillus cereus.
Nature, 338, 1989
|
|
2CZY
 
 | | Solution structure of the NRSF/REST-mSin3B PAH1 complex | | 分子名称: | Paired amphipathic helix protein Sin3b, transcription factor REST (version 3) | | 著者 | Nomura, M, Uda-Tochio, H, Murai, K, Mori, N, Nishimura, Y. | | 登録日 | 2005-07-20 | | 公開日 | 2005-12-20 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Neural Repressor NRSF/REST Binds the PAH1 Domain of the Sin3 Corepressor by Using its Distinct Short Hydrophobic Helix
J.Mol.Biol., 354, 2005
|
|
