5CN9
 
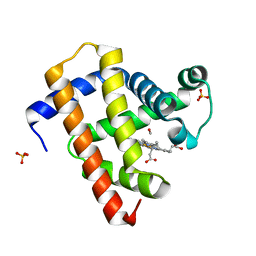 | | Ultrafast dynamics in myoglobin: 0.4 ps time delay | | 分子名称: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
8AXE
 
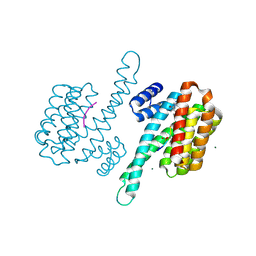 | | Small molecule stabilizer for ERalpha and 14-3-3 (1074210) | | 分子名称: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[2-[1-[2-(4-chloranylphenoxy)-2-methyl-propanoyl]piperidin-4-yl]ethyl]ethanamide, Estrogen receptor, ... | | 著者 | Konstantinidou, M, Visser, E.J, Vandenboorn, E.M.F, Sheng, C, Jaishankar, P, Overmans, M.J.A.M, Dutta, S, Neitz, J, Renslo, A, Ottmann, C, Brunsveld, L, Arkin, M. | | 登録日 | 2022-08-31 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
5CNG
 
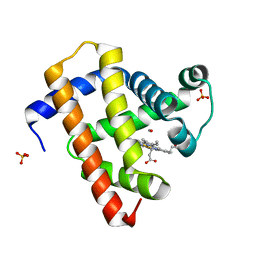 | | ultrafast dynamics in myoglobin: 150 ps time delay | | 分子名称: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
5NN6
 
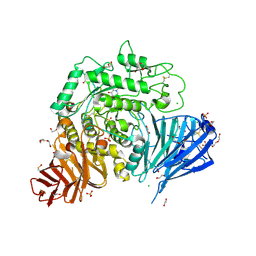 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with N-hydroxyethyl-1-deoxynojirimycin | | 分子名称: | (2R,3R,4R,5S)-1-(2-hydroxyethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Roig-Zamboni, V, Cobucci-Ponzano, B, Iacono, R, Ferrara, M.C, Germany, S, Parenti, G, Bourne, Y, Moracci, M. | | 登録日 | 2017-04-08 | | 公開日 | 2017-10-25 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of human lysosomal acid alpha-glucosidase-a guide for the treatment of Pompe disease.
Nat Commun, 8, 2017
|
|
8E76
 
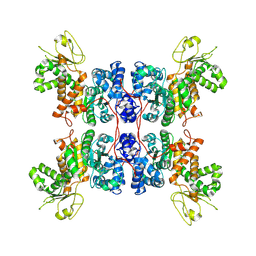 | | Cryo-EM structure of Apo form ME3 | | 分子名称: | NADP-dependent malic enzyme, mitochondrial | | 著者 | Yu, X, Grell, T.A.J, Shaffer, P.L, Steele, R, Sharma, S, Thompson, A.A, Tresadern, G, Ortiz-Meoz, R.F, Mason, M, Gomez-Tamayo, J.C, Riley, D, Wagner, M.V, Wadia, J. | | 登録日 | 2022-08-23 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.51 Å) | | 主引用文献 | Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.
Heliyon, 8, 2022
|
|
1U4J
 
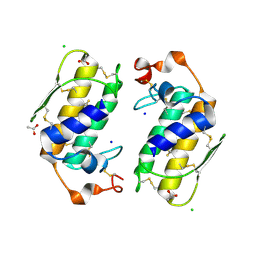 | | Crystal structure of a carbohydrate induced dimer of group I phospholipase A2 from Bungarus caeruleus at 2.1 A resolution | | 分子名称: | ACETIC ACID, CHLORIDE ION, SODIUM ION, ... | | 著者 | Singh, G, Gourinath, S, Sharma, S, Bhanumathi, S, Betzel, C, Srinivasan, A, Singh, T.P. | | 登録日 | 2004-07-26 | | 公開日 | 2004-08-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Crystal structure of a carbohydrate induced homodimer of phospholipase A(2) from Bungarus caeruleus at 2.1A resolution
J.Struct.Biol., 149, 2005
|
|
4QRQ
 
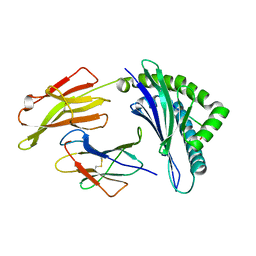 | | Crystal Structure of HLA B*0801 in complex with HSKKKCDEL | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-8 alpha chain, ... | | 著者 | Gras, S, Berry, R, Lucet, I.S, Rossjohn, J. | | 登録日 | 2014-07-02 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | An Extensive Antigenic Footprint Underpins Immunodominant TCR Adaptability against a Hypervariable Viral Determinant.
J.Immunol., 193, 2014
|
|
8E8O
 
 | | Cryo-EM structure of human ME3 in the presence of citrate | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent malic enzyme, mitochondrial | | 著者 | Yu, X, Grell, T.A.J, Shaffer, P.L, Steele, R, Sharma, S, Thompson, A.A, Tresadern, G, Ortiz-Meoz, R.F, Mason, M, Gomez-Tamayo, J.C, Riley, D, Wagner, M.V, Wadia, J. | | 登録日 | 2022-08-25 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.
Heliyon, 8, 2022
|
|
8HB1
 
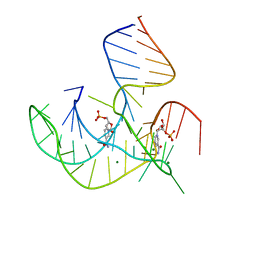 | | Crystal structure of NAD-II riboswitch (two strands) with NMN | | 分子名称: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, MAGNESIUM ION, RNA (30-MER), ... | | 著者 | Peng, X, Lilley, D.M.J, Huang, L. | | 登録日 | 2022-10-27 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Nucleic Acids Res., 51, 2023
|
|
8K9R
 
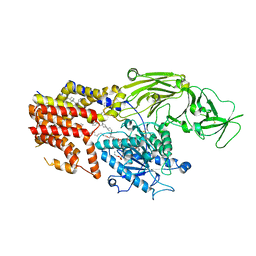 | | Cryo EM structure of the products-bound PGAP1(Bst1)-H443N from Chaetomium thermophilum | | 分子名称: | 2-amino-2-deoxy-alpha-D-glucopyranose, 2-azanylethyl [(2R,3S,4S,5S,6S)-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl hydrogen phosphate, 2-azanylethyl [(2~{S},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl] hydrogen phosphate, ... | | 著者 | Li, T, Hong, J, Qu, Q, Li, D. | | 登録日 | 2023-08-01 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis.
Nat Commun, 15, 2024
|
|
8HBA
 
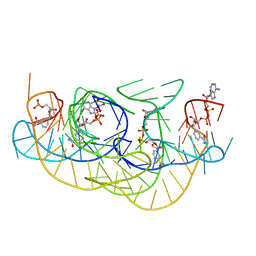 | |
8K9Q
 
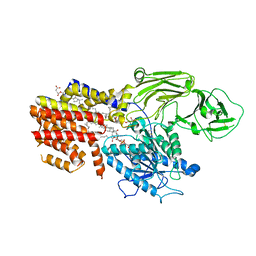 | | Cryo-EM structure of the GPI inositol-deacylase (PGAP1/Bst1) from Chaetomium thermophilum | | 分子名称: | (2~{S})-2-azanyl-3-[[(2~{R})-3-hexadecanoyloxy-2-[(~{Z})-octadec-9-enoyl]oxy-propoxy]-oxidanyl-phosphoryl]oxy-propanoic acid, CHOLESTEROL HEMISUCCINATE, GPI inositol-deacylase,fused thermostable green fluorescent protein | | 著者 | Hong, J, Li, T, Qu, Q, Li, D. | | 登録日 | 2023-08-01 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (2.84 Å) | | 主引用文献 | Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis.
Nat Commun, 15, 2024
|
|
8HB3
 
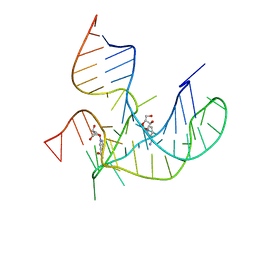 | |
8K9T
 
 | | Cryo-EM structure of the products-bound PGAP1(Bst1)-S327A from Chaetonium thermophilum | | 分子名称: | 2-amino-2-deoxy-alpha-D-glucopyranose, 2-azanylethyl [(2R,3S,4S,5S,6S)-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl hydrogen phosphate, 2-azanylethyl [(2~{S},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl] hydrogen phosphate, ... | | 著者 | Li, T, Hong, J, Qu, Q, Li, D. | | 登録日 | 2023-08-01 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis.
Nat Commun, 15, 2024
|
|
8HB8
 
 | |
8E78
 
 | | Cryo-EM structure of human ME3 in the presence of citrate | | 分子名称: | NADP-dependent malic enzyme, mitochondrial | | 著者 | Yu, X, Grell, T.A.J, Shaffer, P.L, Steele, R, Sharma, S, Thompson, A.A, Tresadern, G, Ortiz-Meoz, R.F, Mason, M, Gomez-Tamayo, J.C, Riley, D, Wagner, M.V, Wadia, J. | | 登録日 | 2022-08-23 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.
Heliyon, 8, 2022
|
|
4JC3
 
 | |
5TZ1
 
 | | Crystal structure of sterol 14-alpha demethylase (CYP51) from Candida albicans in complex with the tetrazole-based antifungal drug candidate VT1161 (VT1) | | 分子名称: | (R)-2-(2,4-Difluorophenyl)-1,1-difluoro-3-(1H-tetrazol-1-yl)-1-(5-(4-(2,2,2-trifluoroethoxy)phenyl)pyridin-2-yl)propan-2-ol, PROTOPORPHYRIN IX CONTAINING FE, Sterol 14-alpha demethylase | | 著者 | Hargrove, T, Wawrzak, Z, Lepesheva, G. | | 登録日 | 2016-11-21 | | 公開日 | 2017-03-15 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural analyses of Candida albicans sterol 14 alpha-demethylase complexed with azole drugs address the molecular basis of azole-mediated inhibition of fungal sterol biosynthesis.
J. Biol. Chem., 292, 2017
|
|
1CB2
 
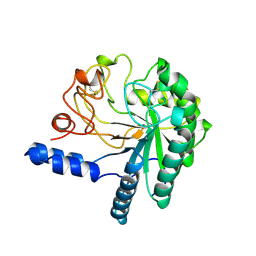 | | CELLOBIOHYDROLASE II, CATALYTIC DOMAIN, MUTANT Y169F | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE II, alpha-D-mannopyranose | | 著者 | Kleywegt, G.J, Szardenings, M, Jones, T.A. | | 登録日 | 1995-11-25 | | 公開日 | 1996-10-14 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The active site of Trichoderma reesei cellobiohydrolase II: the role of tyrosine 169.
Protein Eng., 9, 1996
|
|
1UAG
 
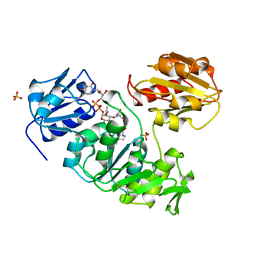 | |
4J80
 
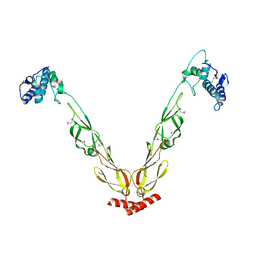 | | Thermus thermophilus DnaJ | | 分子名称: | Chaperone protein DnaJ 2 | | 著者 | Barends, T.R.M, Brosi, R.W, Steinmetz, A, Scherer, A, Hartmann, E, Eschenbach, J, Lorenz, T, Seidel, R, Shoeman, R, Zimmermann, S, Bittl, R, Schlichting, I, Reinstein, J. | | 登録日 | 2013-02-14 | | 公開日 | 2013-07-31 | | 最終更新日 | 2013-10-09 | | 実験手法 | EPR (2.9 Å), X-RAY DIFFRACTION | | 主引用文献 | Combining crystallography and EPR: crystal and solution structures of the multidomain cochaperone DnaJ.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5TZY
 
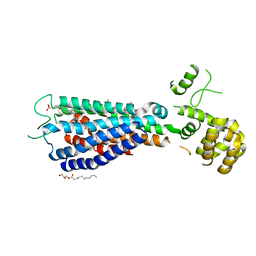 | | GPR40 in complex with AgoPAM AP8 and partial agonist MK-8666 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,3R)-3-cyclopropyl-3-[(2R)-2-(1-{(1S)-1-[5-fluoro-2-(trifluoromethoxy)phenyl]ethyl}piperidin-4-yl)-3,4-dihydro-2H-1-benzopyran-7-yl]-2-methylpropanoic acid, (5aR,6S,6aS)-3-({2',6'-dimethyl-4'-[3-(methylsulfonyl)propoxy][1,1'-biphenyl]-3-yl}methoxy)-5,5a,6,6a-tetrahydrocyclopropa[4,5]cyclopenta[1,2-c]pyridine-6-carboxylic acid, ... | | 著者 | Lu, J, Byrne, N, Patel, S, Sharma, S, Soisson, S.M. | | 登録日 | 2016-11-22 | | 公開日 | 2017-06-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.22 Å) | | 主引用文献 | Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40.
Nat. Struct. Mol. Biol., 24, 2017
|
|
8HJA
 
 | | The crystal structure of syn_CdgR-(c-di-GMP) from Synechocystis sp. PCC 6803 | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), c-di-GMP receptor | | 著者 | Zeng, X, Peng, Y.J. | | 登録日 | 2022-11-22 | | 公開日 | 2023-03-29 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | A c-di-GMP binding effector controls cell size in a cyanobacterium.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ARX
 
 | | Small molecular stabilizer for ERalpha and 14-3-3sigma (1074378) | | 分子名称: | 14-3-3 protein sigma, 2-chloranyl-N-[[1-[1-(4-chloranylphenoxy)cyclopropyl]carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | 著者 | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | 登録日 | 2022-08-17 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8ALV
 
 | | Small molecular stabilizer for ERalpha and 14-3-3 (1076403) | | 分子名称: | 14-3-3 protein sigma, 2-chloranyl-N-[[1-[1-[(4-chlorophenyl)amino]cyclohexyl]carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | 著者 | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | 登録日 | 2022-08-01 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
