6PWB
 
 | | Rigid body fitting of flagellin FlaB, and flagellar coiling proteins, FcpA and FcpB, into a 10 Angstrom structure of the asymmetric flagellar filament purified from Leptospira biflexa Patoc WT cells resolved via subtomogram averaging | | 分子名称: | Flagellar coiling protein A (FcpA), Flagellar coiling protein B (FcpB), Flagellin B1 (FlaB1) | | 著者 | Gibson, K.H, Sindelar, C.V, Trajtenberg, F, Buschiazzo, A, San Martin, F, Mechaly, A. | | 登録日 | 2019-07-22 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (9.83 Å) | | 主引用文献 | An asymmetric sheath controls flagellar supercoiling and motility in the leptospira spirochete.
Elife, 9, 2020
|
|
6W7S
 
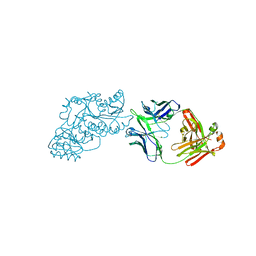 | | Ketoreductase from module 1 of the 6-deoxyerythronolide B synthase (KR1) in complex with antibody fragment (Fab) 2G10 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2G10 (Fab heavy chain), 2G10 (Fab light chain), ... | | 著者 | Cogan, D.P, Mathews, I.I, Khosla, C. | | 登録日 | 2020-03-19 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Antibody Probes of Module 1 of the 6-Deoxyerythronolide B Synthase Reveal an Extended Conformation During Ketoreduction.
J.Am.Chem.Soc., 142, 2020
|
|
8UOK
 
 | | Crystal structure of human NUAK1-MARK3 (7 mutations) kinase domain chimera bound with small molecule inhibitor #31 | | 分子名称: | (6P)-6-[(4R)-imidazo[1,2-a]pyridin-3-yl]-4-(piperidin-4-yl)-2H-pyrido[3,2-b][1,4]oxazin-3(4H)-one, 1,2-ETHANEDIOL, MAP/microtubule affinity-regulating kinase 3 | | 著者 | Delker, S.L, Abendroth, J, Fox III, D.D. | | 登録日 | 2023-10-19 | | 公開日 | 2024-10-23 | | 最終更新日 | 2024-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Discovery of UCB9386: A Potent, Selective, and Brain-Penetrant Nuak1 Inhibitor Suitable for In Vivo Pharmacological Studies.
J.Med.Chem., 67, 2024
|
|
6O97
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with propylamycin and bound to mRNA and A-, P-, and E-site tRNAs at 2.75A resolution | | 分子名称: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxyc yclohexyl 2-amino-2,4-dideoxy-4-propyl-alpha-D-glucopyranoside, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | 著者 | Matsushita, T, Sati, G.C, Kondasinghe, N, Pirrone, M.G, Kato, T, Waduge, P, Kumar, H.S, Sanchon, A.C, Dobosz-Bartoszek, M, Shcherbakov, D, Juhas, M, Hobbie, S.N, Schrepfer, T, Chow, C.S, Polikanov, Y.S, Schacht, J, Vasella, A, Bottger, E.C, Crich, D. | | 登録日 | 2019-03-13 | | 公開日 | 2019-04-17 | | 最終更新日 | 2025-03-19 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Design, Multigram Synthesis, and in Vitro and in Vivo Evaluation of Propylamycin: A Semisynthetic 4,5-Deoxystreptamine Class Aminoglycoside for the Treatment of Drug-Resistant Enterobacteriaceae and Other Gram-Negative Pathogens.
J. Am. Chem. Soc., 141, 2019
|
|
2A68
 
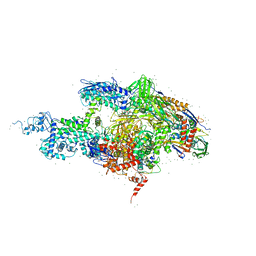 | | Crystal structure of the T. thermophilus RNA polymerase holoenzyme in complex with antibiotic rifabutin | | 分子名称: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | 著者 | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-07-01 | | 公開日 | 2005-09-20 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
5WIS
 
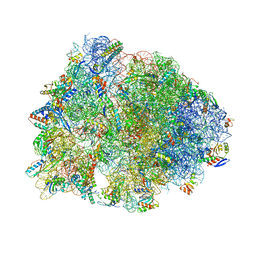 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with methymycin and bound to mRNA and A-, P- and E-site tRNAs at 2.7A resolution | | 分子名称: | (3R,4S,5S,7R,9E,11S,12R)-12-ethyl-11-hydroxy-3,5,7,11-tetramethyl-2,8-dioxooxacyclododec-9-en-4-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 16S Ribosomal RNA, 23S ribosomal RNA, ... | | 著者 | Almutairi, M.M, Svetlov, M.S, Hansen, D.A, Khabibullina, N.F, Klepacki, D, Kang, H.Y, Sherman, D.H, Vazquez-Laslop, N, Polikanov, Y.S, Mankin, A.S. | | 登録日 | 2017-07-20 | | 公開日 | 2018-02-14 | | 最終更新日 | 2025-03-19 | | 実験手法 | X-RAY DIFFRACTION (2.703 Å) | | 主引用文献 | Co-produced natural ketolides methymycin and pikromycin inhibit bacterial growth by preventing synthesis of a limited number of proteins.
Nucleic Acids Res., 45, 2017
|
|
6ORD
 
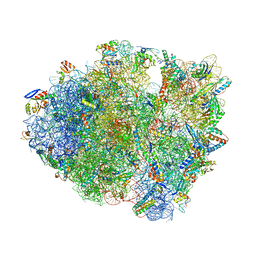 | | Crystal structure of tRNA^ Ala(GGC) U32-A38 bound to cognate 70S A site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Nguyen, H.A, Sunita, S, Dunham, C.M. | | 登録日 | 2019-04-30 | | 公開日 | 2020-06-24 | | 最終更新日 | 2025-03-19 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Disruption of evolutionarily correlated tRNA elements impairs accurate decoding.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8QL3
 
 | | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution: 233 fs structure | | 分子名称: | Azo-Combretastatin A4 (cis), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | 著者 | Weinert, T, Wranik, M, Seidel, H.-P, Church, J, Steinmetz, M.O, Schapiro, I, Standfuss, J. | | 登録日 | 2023-09-19 | | 公開日 | 2024-10-02 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution
To Be Published
|
|
8QL4
 
 | | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution: 349 fs structure | | 分子名称: | Azo-Combretastatin A4 (cis), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | 著者 | Weinert, T, Wranik, M, Seidel, H.-P, Church, J, Steinmetz, M.O, Schapiro, I, Standfuss, J. | | 登録日 | 2023-09-19 | | 公開日 | 2024-10-02 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution
To Be Published
|
|
6OF6
 
 | | Crystal structure of tRNA^ Ala(GGC) bound to cognate 70S A-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Nguyen, H.A, Sunita, S, Dunham, C.M. | | 登録日 | 2019-03-28 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Disruption of evolutionarily correlated tRNA elements impairs accurate decoding.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OPE
 
 | | Crystal structure of tRNA^ Ala(GGC) U32-A38 bound to near-cognate 70S A site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Nguyen, H.A, Sunita, S, Dunham, C.M. | | 登録日 | 2019-04-24 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Disruption of evolutionarily correlated tRNA elements impairs accurate decoding.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OJ2
 
 | | Crystal structure of tRNA^ Ala(GGC) bound to the near-cognate 70S A-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Nguyen, H.A, Sunita, S, Dunham, C.M. | | 登録日 | 2019-04-10 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Disruption of evolutionarily correlated tRNA elements impairs accurate decoding.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8FON
 
 | | Crystal structure of tRNA^Lys(SUU) bound to AUA codon in the ribosomal P site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Nguyen, H.A, Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | 登録日 | 2023-01-02 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (3.64 Å) | | 主引用文献 | Structural basis for reduced ribosomal A-site fidelity in response to P-site codon-anticodon mismatches.
J.Biol.Chem., 299, 2023
|
|
8FOM
 
 | | Crystal structure of tRNA^Lys(SUU) bound to UAA codon in the ribosomal P site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Nguyen, H.A, Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | 登録日 | 2023-01-02 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (3.58 Å) | | 主引用文献 | Structural basis for reduced ribosomal A-site fidelity in response to P-site codon-anticodon mismatches.
J.Biol.Chem., 299, 2023
|
|
2A69
 
 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with antibiotic rifapentin | | 分子名称: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | 著者 | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-07-02 | | 公開日 | 2005-09-20 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
6BY1
 
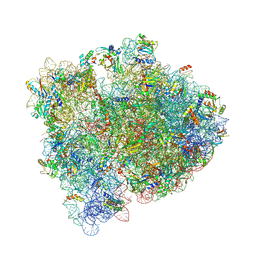 | | E. coli pH03H9 complex | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Amiri, H, Noller, H.F. | | 登録日 | 2017-12-19 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (3.94 Å) | | 主引用文献 | Structural evidence for product stabilization by the ribosomal mRNA helicase.
Rna, 25, 2019
|
|
8QEA
 
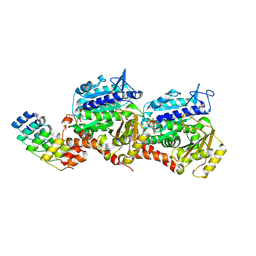 | | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution: 96 fs structure | | 分子名称: | Azo-Combretastatin A4 (cis), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | 著者 | Weinert, T, Wranik, M, Seidel, H.-P, Church, J, Steinmetz, M.O, Schapiro, I, Standfuss, J. | | 登録日 | 2023-08-31 | | 公開日 | 2024-09-11 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution
To Be Published
|
|
8RXH
 
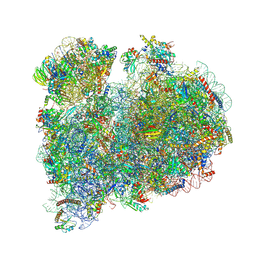 | | CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME WITH A/P/E-site tRNA AND mRNA : PARENTAL STRAIN | | 分子名称: | (2S)-2-[2-[4-[[(2R,3S,4S)-3-acetyloxy-4-oxidanyl-pyrrolidin-2-yl]methyl]phenoxy]ethanoylamino]-6-azanyl-hexanoic acid, 40S ribosomal protein S12, 40S ribosomal protein S14, ... | | 著者 | Rajan, K.S, Yonath, A. | | 登録日 | 2024-02-07 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Structural and mechanistic insights into the function of Leishmania ribosome lacking a single pseudouridine modification.
Cell Rep, 43, 2024
|
|
7LZE
 
 | | Cryo-EM Structure of disulfide stabilized HMPV F v4-B | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0 | | 著者 | Gorman, J, Kwong, P.D. | | 登録日 | 2021-03-09 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.23 Å) | | 主引用文献 | Interprotomer disulfide-stabilized variants of the human metapneumovirus fusion glycoprotein induce high titer-neutralizing responses.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
9E78
 
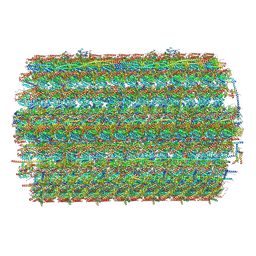 | | 48-nm repeat of the Leishmania tarentolae doublet microtubule | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, ArcMAP1, ArcMAP2, ... | | 著者 | Doran, M.H, Ren, P, Hoog, J.L, Brown, A. | | 登録日 | 2024-11-01 | | 公開日 | 2025-03-12 | | 最終更新日 | 2025-03-26 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Evolutionary adaptations of doublet microtubules in trypanosomatid parasites.
Science, 387, 2025
|
|
7CRL
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 50 ps after light activation | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-13 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRY
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (6.49 mJ/mm2) | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRK
 
 | | 2ps Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-13 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRX
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (2.63mJ/mm2) | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRT
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (0.17mJ/mm2) | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
