3W38
 
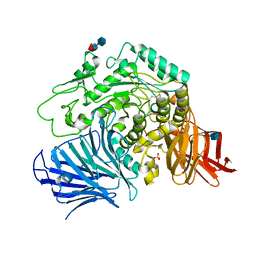 | | Sugar beet alpha-glucosidase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-glucosidase, SULFATE ION, ... | | 著者 | Tagami, T, Yamashita, K, Okuyama, M, Mori, H, Yao, M, Kimura, A. | | 登録日 | 2012-12-13 | | 公開日 | 2013-05-29 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Molecular basis for the recognition of long-chain substrates by plant & alpha-glucosidase
J.Biol.Chem., 288, 2013
|
|
8A8J
 
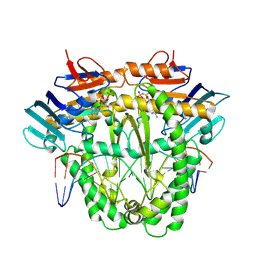 | | Complex of RecF and DNA from Thermus thermophilus. | | 分子名称: | DNA replication and repair protein RecF, MAGNESIUM ION, Oligo1, ... | | 著者 | Nirwal, S, Czarnocki-Cieciura, M, Chaudhary, A, Zajko, W, Skowronek, K, Chamera, S, Figiel, M, Nowotny, M. | | 登録日 | 2022-06-23 | | 公開日 | 2023-04-26 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1SHK
 
 | |
1S30
 
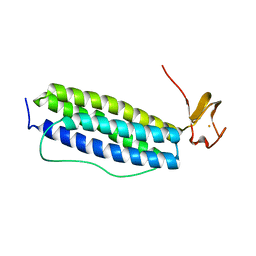 | | X-ray crystal structure of Desulfovibrio vulgaris Rubrerythrin with displacement of iron by zinc at the diiron Site | | 分子名称: | FE (III) ION, Rubrerythrin, ZINC ION | | 著者 | Jin, S, Kurtz Jr, D.M, Liu, Z.-J, Rose, J, Wang, B.-C. | | 登録日 | 2004-01-12 | | 公開日 | 2004-06-22 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Displacement of iron by zinc at the diiron site of Desulfovibrio vulgaris rubrerythrin: X-ray crystal structure and anomalous scattering analysis
J.Inorg.Biochem., 98, 2004
|
|
4XXF
 
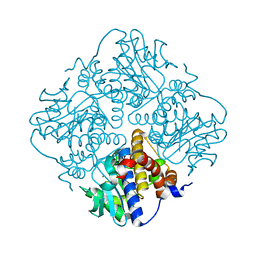 | | L-fuculose 1-phosphate aldolase from Glaciozyma antarctica PI12 | | 分子名称: | Fuculose-1-phosphate aldolase, ZINC ION | | 著者 | Jaafar, N.R, Abu Bakar, F.D, Abdul Murad, A.M, Illias, R, Littler, D, Beddoe, T, Rossjohn, J, Mahadi, M.N. | | 登録日 | 2015-01-30 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Crystal structure of fuculose aldolase from the Antarctic psychrophilic yeast Glaciozyma antarctica PI12.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
1R81
 
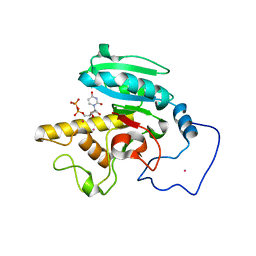 | | Glycosyltransferase A in complex with 3-amino-acceptor analog inhibitor and uridine diphosphate-N-acetyl-galactose | | 分子名称: | Glycoprotein-fucosylgalactoside alpha-N-acetylgalactosaminyltransferase, MERCURY (II) ION, URIDINE-DIPHOSPHATE-N-ACETYLGALACTOSAMINE, ... | | 著者 | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | 登録日 | 2003-10-22 | | 公開日 | 2004-02-10 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
2C1E
 
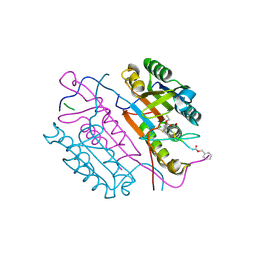 | | Crystal structures of caspase-3 in complex with aza-peptide Michael acceptor inhibitors. | | 分子名称: | AZA-PEPTIDE INHIBITOR (5S, 8R, 11S)-8-(2-CARBOXYETHYL)-5-(CARBOXYMETHYL)-14-(4-ETHOXY-4-OXOBUTANOYL)-11-(1-METHYLETHYL)-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,13,14-PENTAAZAHEXADECAN -16-OIC ACID, ... | | 著者 | Grutter, M.G. | | 登録日 | 2005-09-14 | | 公開日 | 2006-09-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Design, Synthesis, and Evaluation of Aza-Peptide Michael Acceptors as Selective and Potent Inhibitors of Caspases-2, -3, -6, -7, -8, -9, and - 10.
J.Med.Chem., 49, 2006
|
|
2C2M
 
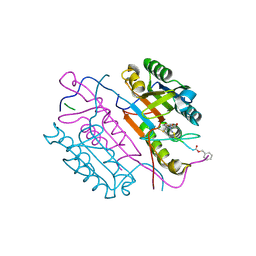 | | Crystal structures of caspase-3 in complex with aza-peptide Michael acceptor inhibitors. | | 分子名称: | AZA-PEPTIDE INHIBITOR (5S, 8R, 11S)-14-[4-(BENZYLOXY)-4-OXOBUTANOYL]-8-(2-CARBOXYETHYL)-5-(CARBOXYMETHYL)-11-(1-METHYLETHYL)-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,13,14 -PENTAAZAHEXADECAN-16-OIC ACID, ... | | 著者 | Ganesan, R, Jelakovic, S, Ekici, O.D, Li, Z.Z, James, K.E, Asgian, J.L, Campbell, A, Mikolajczyk, J, Salvesen, G.S, Gruetter, M.G, Powers, J.C. | | 登録日 | 2005-09-29 | | 公開日 | 2006-09-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Design, Synthesis, and Evaluation of Aza-Peptide Michael Acceptors as Selective and Potent Inhibitors of Caspases-2, -3, -6, -7, -8, -9, and - 10.
J.Med.Chem., 49, 2006
|
|
1R1V
 
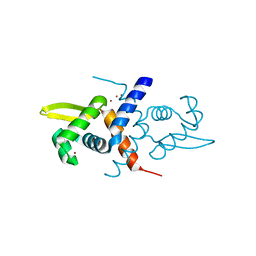 | | Crystal structure of the metal-sensing transcriptional repressor CzrA from Staphylococcus aureus in the Zn2-form | | 分子名称: | ZINC ION, repressor protein | | 著者 | Eicken, C, Pennella, M.A, Chen, X, Koshlap, K.M, VanZile, M.L, Sacchettini, J.C, Giedroc, D.P. | | 登録日 | 2003-09-25 | | 公開日 | 2004-05-18 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins.
J.Mol.Biol., 333, 2003
|
|
5EZU
 
 | | Crystal structure of the N-terminal domain of vaccinia virus immunomodulator A46 in complex with myristic acid. | | 分子名称: | MYRISTIC ACID, Protein A46 | | 著者 | Fedosyuk, S, Bezerra, G.A, Sammito, M, Uson, I, Skern, T. | | 登録日 | 2015-11-26 | | 公開日 | 2016-11-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Vaccinia Virus Immunomodulator A46: A Lipid and Protein-Binding Scaffold for Sequestering Host TIR-Domain Proteins.
PLoS Pathog., 12, 2016
|
|
2DU6
 
 | |
6F62
 
 | |
8JM9
 
 | |
8JMI
 
 | |
8JMA
 
 | |
8JME
 
 | |
8JMH
 
 | |
1RNL
 
 | | THE NITRATE/NITRITE RESPONSE REGULATOR PROTEIN NARL FROM NARL | | 分子名称: | GLYCEROL, NITRATE/NITRITE RESPONSE REGULATOR PROTEIN NARL, PLATINUM (II) ION | | 著者 | Baikalov, I, Schroder, I, Kaczor-Grzeskowiak, M, Grzeskowiak, K, Gunsalus, R.P, Dickerson, R.E. | | 登録日 | 1996-04-17 | | 公開日 | 1996-11-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of the Escherichia coli response regulator NarL.
Biochemistry, 35, 1996
|
|
3F5K
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | 著者 | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | 登録日 | 2008-11-03 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
1S2Z
 
 | | X-ray crystal structure of Desulfovibrio vulgaris Rubrerythrin with displacement of iron by zinc at the diiron Site | | 分子名称: | FE (III) ION, Rubrerythrin, ZINC ION | | 著者 | Jin, S, Kurtz Jr, D.M, Liu, Z.-J, Rose, J, Wang, B.-C. | | 登録日 | 2004-01-12 | | 公開日 | 2004-06-22 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Displacement of iron by zinc at the diiron site of Desulfovibrio vulgaris rubrerythrin: X-ray crystal structure and anomalous scattering analysis
J.Inorg.Biochem., 98, 2004
|
|
4D5I
 
 | | Hypocrea jecorina cellobiohydrolase Cel7A E212Q soaked with xylotriose. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLULOSE 1,4-BETA-CELLOBIOSIDASE, COBALT (II) ION, ... | | 著者 | Momeni, M.H, Ubhayasekera, W, Stahlberg, J, Hansson, H. | | 登録日 | 2014-11-05 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Structural Insights Into the Inhibition of Cellobiohydrolase Cel7A by Xylooligosaccharides.
FEBS J., 282, 2015
|
|
4J2C
 
 | | GARP-SNARE Interaction | | 分子名称: | Syntaxin-6, Vacuolar protein sorting-associated protein 51 homolog | | 著者 | Abascal-Palacios, G, Schindler, C, Rojas, A.L, Bonifacino, J.S, Hierro, A. | | 登録日 | 2013-02-04 | | 公開日 | 2013-12-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Structural basis for the interaction of the Golgi-Associated Retrograde Protein Complex with the t-SNARE Syntaxin 6.
Structure, 21, 2013
|
|
4NWP
 
 | | Computationally Designed Two-Component Self-Assembling Tetrahedral Cage, T33-21, Crystallized in Space Group R32 | | 分子名称: | AMMONIUM ION, Putative uncharacterized protein, SULFATE ION, ... | | 著者 | McNamara, D.E, King, N.P, Bale, J.B, Sheffler, W, Baker, D, Yeates, T.O. | | 登録日 | 2013-12-06 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Accurate design of co-assembling multi-component protein nanomaterials.
Nature, 510, 2014
|
|
6F5X
 
 | |
2N0J
 
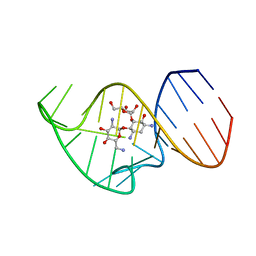 | | Solution NMR Structure of the 27 nucleotide engineered neomycin sensing riboswitch RNA-ribostamycin complex | | 分子名称: | RIBOSTAMYCIN, RNA_(27-MER) | | 著者 | Duchardt-Ferner, E, Gottstein-Schmidtke, S.R, Weigand, J.E, Ohlenschlaeger, O.E, Wurm, J, Hammann, C, Suess, B, Woehnert, J. | | 登録日 | 2015-03-09 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | What a Difference an OH Makes: Conformational Dynamics as the Basis for the Ligand Specificity of the Neomycin-Sensing Riboswitch.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
