4FIX
 
 | | Crystal Structure of GlfT2 | | 分子名称: | BETA-MERCAPTOETHANOL, GLYCEROL, MANGANESE (II) ION, ... | | 著者 | Wheatley, R.W, Zheng, R.B, Lowary, T.L, Ng, K.K.S. | | 登録日 | 2012-06-11 | | 公開日 | 2012-06-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Tetrameric Structure of the GlfT2 Galactofuranosyltransferase Reveals a Scaffold for the Assembly of Mycobacterial Arabinogalactan.
J.Biol.Chem., 287, 2012
|
|
2QRX
 
 | |
2QVA
 
 | |
8XBT
 
 | | The cryo-EM structure of the octameric RAD51 ring bound to the nucleosome with the linker DNA binding | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.12 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8U3Y
 
 | | SpG Cas9 with NGG PAM DNA target | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(P*CP*GP*TP*TP*TP*GP*TP*AP*CP*TP*CP*CP*AP*GP*CP*G)-3'), DNA (5'-D(P*TP*CP*TP*CP*AP*TP*CP*TP*TP*TP*AP*TP*GP*CP*GP*TP*C)-3'), ... | | 著者 | Bravo, J.P.K, Hibshman, G.N, Taylor, D.W. | | 登録日 | 2023-09-08 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
8TZZ
 
 | | SpG Cas9 with NGC PAM DNA target | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(P*CP*GP*TP*TP*TP*GP*TP*AP*CP*TP*GP*CP*AP*GP*CP*G)-3'), DNA (5'-D(P*TP*CP*TP*CP*AP*TP*CP*TP*TP*TP*AP*TP*GP*CP*GP*TP*C)-3'), ... | | 著者 | Bravo, J.P.K, Hibshman, G.N, Taylor, D.W. | | 登録日 | 2023-08-28 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.56 Å) | | 主引用文献 | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
8W7P
 
 | | Extracellular domain of a sensor histidine kinase | | 分子名称: | Extracellular domain of a sensor histidine kinase NagS, GLYCEROL | | 著者 | Itoh, T, Ogawa, T, Hibi, T, Kimoto, H. | | 登録日 | 2023-08-31 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Characterization of the extracellular domain of sensor histidine kinase NagS from Paenibacillus sp. str. FPU-7: nagS interacts with oligosaccharide binding protein NagB1 in complexes with N, N'-diacetylchitobiose.
Biosci.Biotechnol.Biochem., 88, 2024
|
|
8XGV
 
 | | Optimization Efforts for Identification of Novel Highly Potent Keap1-Nrf2 Protein-Protein Interaction (PPI) Inhibitors | | 分子名称: | (2~{R},3~{S})-3-[[(2~{S})-2-[4-[(3-ethoxypyridin-2-yl)methyl]phenyl]-2-fluoranyl-ethanoyl]amino]-2-methyl-3-(4-methylphenyl)propanoic acid, ACETATE ION, Kelch-like ECH-associated protein 1, ... | | 著者 | Otake, K, Hara, Y, Ubukata, M, Inoue, M, Nagahashi, N, Motoda, D, Ogawa, N, Hantani, Y, Hantani, R, Adachi, T, Nomura, A, Yamaguchi, K, Maekawa, M, Mamada, H, Motomura, T, Sato, M, Harada, K. | | 登録日 | 2023-12-15 | | 公開日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Optimization Efforts for Identification of Novel Highly Potent Keap1-Nrf2 Protein-Protein Interaction Inhibitors.
J.Med.Chem., 67, 2024
|
|
8WM0
 
 | | Crystal structure of TNIK-thiopeptide wTP3 complex | | 分子名称: | ADENOSINE, THIOPEPTIDE wTP3, TRAF2 and NCK-interacting protein kinase | | 著者 | Hamada, K, Kobayashi, S, Vinogradov, A.A, Zhang, Y, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | 登録日 | 2023-10-01 | | 公開日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A Compact Reprogrammed Genetic Code for De Novo Discovery of Proteolytically Stable Thiopeptides.
J.Am.Chem.Soc., 2024
|
|
8XGK
 
 | | Optimization Efforts for Identification of Novel Highly Potent Keap1-Nrf2 Protein-Protein Interaction Ihhibitors | | 分子名称: | (2~{R},3~{S})-3-[[(2~{S})-2-(4-chlorophenyl)-2-fluoranyl-ethanoyl]amino]-3-[3-(2-cyano-2-methyl-propoxy)-4-methyl-phenyl]-2-methyl-propanoic acid, ACETATE ION, Kelch-like ECH-associated protein 1, ... | | 著者 | Otake, K, Hara, Y, Ubukata, M, Inoue, M, Nagahashi, N, Motoda, D, Ogawa, N, Hantani, Y, Hantani, R, Adachi, T, Nomura, A, Yamaguchi, K, Maekawa, M, Mamada, H, Motomura, T, Sato, M, Harada, K. | | 登録日 | 2023-12-15 | | 公開日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Optimization Efforts for Identification of Novel Highly Potent Keap1-Nrf2 Protein-Protein Interaction Inhibitors.
J.Med.Chem., 67, 2024
|
|
8XBV
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the sticky end of the nucleosome | | 分子名称: | DNA (5'-D(P*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*A)-3'), DNA (5'-D(P*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*G)-3'), DNA repair protein RAD51 homolog 1 | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.61 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBW
 
 | | The cryo-EM structure of the RAD51 N-terminal lobe domain bound to the histone H4 tail of the nucleosome | | 分子名称: | DNA (5'-D(P*AP*CP*CP*GP*CP*TP*TP*AP*AP*AP*CP*GP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*CP*GP*TP*TP*TP*AP*AP*GP*CP*GP*GP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBY
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the blunt end of the nucleosome | | 分子名称: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.8 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBU
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome with the linker DNA binding | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.24 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBX
 
 | | The cryo-EM structure of the RAD51 L2 loop bound to the linker DNA with the blunt end of the nucleosome | | 分子名称: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (4.36 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
7KKJ
 
 | | Structure of anti-SARS-CoV-2 Spike nanobody mNb6 | | 分子名称: | CHLORIDE ION, SULFATE ION, Synthetic nanobody mNb6 | | 著者 | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | 登録日 | 2020-10-27 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKK
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody Nb6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | 登録日 | 2020-10-27 | | 公開日 | 2020-11-11 | | 最終更新日 | 2021-04-21 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKL
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody mNb6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | 登録日 | 2020-10-27 | | 公開日 | 2020-11-11 | | 最終更新日 | 2021-04-21 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KTR
 
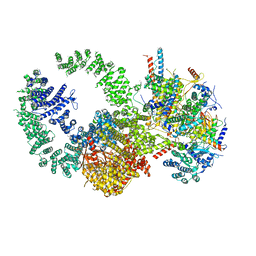 | | Cryo-EM structure of the human SAGA coactivator complex (TRRAP, core) | | 分子名称: | Ataxin-7, INOSITOL HEXAKISPHOSPHATE, Isoform 3 of Transcription factor SPT20 homolog, ... | | 著者 | Herbst, D.A, Esbin, M.N, Nogales, E. | | 登録日 | 2020-11-24 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Structure of the human SAGA coactivator complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KTS
 
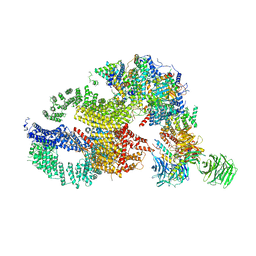 | | Negative stain EM structure of the human SAGA coactivator complex (TRRAP, core, splicing module) | | 分子名称: | Ataxin-7, Isoform 3 of Transcription factor SPT20 homolog, STAGA complex 65 subunit gamma, ... | | 著者 | Herbst, D.A, Esbin, M.N, Nogales, E. | | 登録日 | 2020-11-24 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (19.09 Å) | | 主引用文献 | Structure of the human SAGA coactivator complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1FBT
 
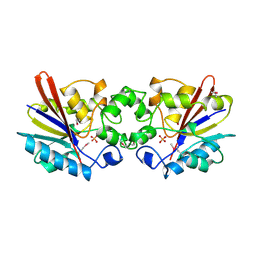 | | THE BISPHOSPHATASE DOMAIN OF THE BIFUNCTIONAL RAT LIVER 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE | | 分子名称: | FRUCTOSE-2,6-BISPHOSPHATASE, PHOSPHATE ION | | 著者 | Lee, Y.-H, Ogata, C, Pflugrath, J.W, Levitt, D.G, Sarma, R, Banaszak, L.J, Pilkis, S.J. | | 登録日 | 1996-03-08 | | 公開日 | 1997-07-23 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the rat liver fructose-2,6-bisphosphatase based on selenomethionine multiwavelength anomalous dispersion phases.
Biochemistry, 35, 1996
|
|
7LYS
 
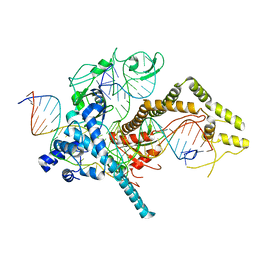 | | Cryo-EM structure of CasPhi-2 (Cas12j) bound to crRNA and DNA | | 分子名称: | CasPhi-2, NTS-DNA, TS-DNA, ... | | 著者 | Pausch, P, Soczek, K, Nogales, E, Doudna, J. | | 登録日 | 2021-03-08 | | 公開日 | 2021-08-04 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | DNA interference states of the hypercompact CRISPR-Cas Phi effector.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LYT
 
 | | Cryo-EM structure of CasPhi-2 (Cas12j) bound to crRNA and Phosphorothioate-DNA | | 分子名称: | CasPhi, MAGNESIUM ION, NTS-DNA*, ... | | 著者 | Pausch, P, Soczek, K, Nogales, E, Doudna, J. | | 登録日 | 2021-03-08 | | 公開日 | 2021-08-04 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | DNA interference states of the hypercompact CRISPR-Cas Phi effector.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1GC5
 
 | | CRYSTAL STRUCTURE OF A NOVEL ADP-DEPENDENT GLUCOKINASE FROM THERMOCOCCUS LITORALIS | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADP-DEPENDENT GLUCOKINASE | | 著者 | Ito, S, Fushinobu, S, Yoshioka, I, Koga, S, Matsuzawa, H, Wakagi, T. | | 登録日 | 2000-07-20 | | 公開日 | 2001-07-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Basis for the ADP-Specificity of a Novel Glucokinase from a Hyperthermophilic Archaeon
Structure, 9, 2001
|
|
1GIR
 
 | | CRYSTAL STRUCTURE OF THE ENZYMATIC COMPONET OF IOTA-TOXIN FROM CLOSTRIDIUM PERFRINGENS WITH NADPH | | 分子名称: | IOTA TOXIN COMPONENT IA, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Tsuge, H, Nagahama, M, Nishimura, H, Hisatsune, J, Sakaguchi, Y, Itogawa, Y, Katunuma, N, Sakurai, J. | | 登録日 | 2001-03-12 | | 公開日 | 2003-01-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure and Site-directed Mutagenesis of Enzymatic Components from Clostridium perfringens Iota-toxin
J.MOL.BIOL., 325, 2003
|
|
