1UVL
 
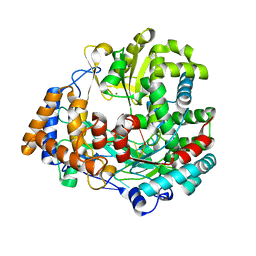 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 with 5nt RNA. Conformation B | | 分子名称: | 5'-R(*UP*UP*UP*CP*CP)-3', MANGANESE (II) ION, RNA-directed RNA polymerase | | 著者 | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | 登録日 | 2004-01-21 | | 公開日 | 2004-02-26 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
1TXI
 
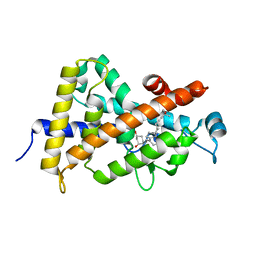 | | Crystal structure of the vdr ligand binding domain complexed to TX522 | | 分子名称: | (1R,3R)-5-((Z)-2-((1R,7AS)-HEXAHYDRO-1-((S)-6-HYDROXY-6-METHYLHEPT-4-YN-2-YL)-7A-METHYL-1H-INDEN-4(7AH)-YLIDENE)ETHYLIDENE)CYCLOHEXANE-1,3-DIOL, Vitamin D receptor | | 著者 | Moras, D, Rochel, N. | | 登録日 | 2004-07-05 | | 公開日 | 2005-05-10 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Superagonistic Action of 14-epi-Analogs of 1,25-Dihydroxyvitamin D Explained by Vitamin D Receptor-Coactivator Interaction
Mol.Pharmacol., 67, 2005
|
|
1HX7
 
 | |
8Q5I
 
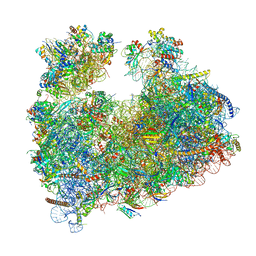 | | Structure of Candida albicans 80S ribosome in complex with cephaeline | | 分子名称: | 18S ribosomal RNA, 25S rRNA, 40S ribosomal protein S0, ... | | 著者 | Kolosova, O, Zgadzay, Y, Stetsenko, A, Atamas, A, Guskov, A, Yusupov, M. | | 登録日 | 2023-08-09 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (2.45 Å) | | 主引用文献 | Structural characterization of cephaeline binding to the eukaryotic ribosome using Cryo-Electron Microscopy
Biopolym Cell, 2023
|
|
8J9T
 
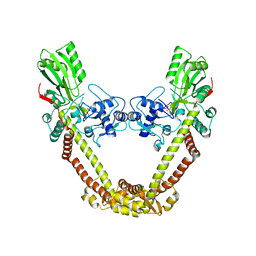 | | Crystal Structure of GyraseA N-terminal at 2.43A Resolution | | 分子名称: | CARBONATE ION, DNA gyrase subunit A | | 著者 | Salman, M, Sachdeva, E, Das, U, Singh, T.P, Ethayathullah, A.S, Kaur, P. | | 登録日 | 2023-05-04 | | 公開日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.428 Å) | | 主引用文献 | Crystal Structure of GyraseA N-terminal at 2.43A Resolution
to be published
|
|
7GQU
 
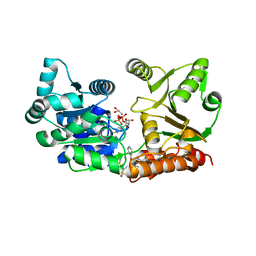 | | Crystal Structure of Werner helicase fragment 517-945 in covalent complex with N-[(E,1S)-1-cyclopropyl-3-methylsulfonylprop-2-enyl]-2-(1,1-difluoroethyl)-4-phenoxypyrimidine-5-carboxamide | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, GLYCEROL, ... | | 著者 | Classen, M, Benz, J, Brugger, D, Tagliente, O, Rudolph, M.G. | | 登録日 | 2023-10-19 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Chemoproteomic discovery of a covalent allosteric inhibitor of WRN helicase.
Nature, 629, 2024
|
|
7GQT
 
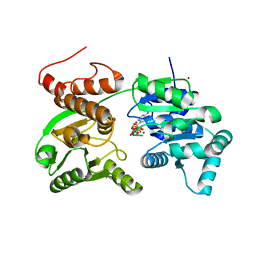 | | Crystal Structure of Werner helicase fragment 517-945 in complex with ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, MAGNESIUM ION, ... | | 著者 | Classen, M, Benz, J, Brugger, D, Rudolph, M.G. | | 登録日 | 2023-10-19 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Chemoproteomic discovery of a covalent allosteric inhibitor of WRN helicase.
Nature, 629, 2024
|
|
6EG7
 
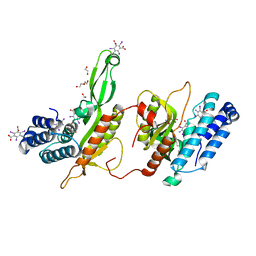 | | BbvCI B2 dimer with I3C clusters | | 分子名称: | 1,2-ETHANEDIOL, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, BbvCI endonuclease subunit 2, ... | | 著者 | Shen, B.W, Stoddard, B.L. | | 登録日 | 2018-08-19 | | 公開日 | 2018-11-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure, subunit organization and behavior of the asymmetric Type IIT restriction endonuclease BbvCI.
Nucleic Acids Res., 47, 2019
|
|
7GQS
 
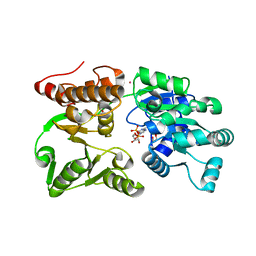 | | Crystal Structure of Werner helicase fragment 517-945 in complex with ADP | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, ... | | 著者 | Classen, M, Benz, J, Brugger, D, Rudolph, M.G. | | 登録日 | 2023-10-19 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Chemoproteomic discovery of a covalent allosteric inhibitor of WRN helicase.
Nature, 629, 2024
|
|
6RVS
 
 | | Atomic structure of the Epstein-Barr portal, structure II | | 分子名称: | Portal protein | | 著者 | Machon, C, Fabrega-Ferrer, M, Zhou, D, Cuervo, A, Carrascosa, J.L, Stuart, D.I, Coll, M. | | 登録日 | 2019-05-31 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.59 Å) | | 主引用文献 | Atomic structure of the Epstein-Barr virus portal.
Nat Commun, 10, 2019
|
|
2LUA
 
 | | Solution structure of CXC domain of MSL2 | | 分子名称: | Protein male-specific lethal-2, ZINC ION | | 著者 | Feng, Y, Ye, K, Zheng, S, Wang, J. | | 登録日 | 2012-06-09 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of MSL2 CXC Domain Reveals an Unusual Zn(3)Cys(9) Cluster and Similarity to Pre-SET Domains of Histone Lysine Methyltransferases.
Plos One, 7, 2012
|
|
4ASN
 
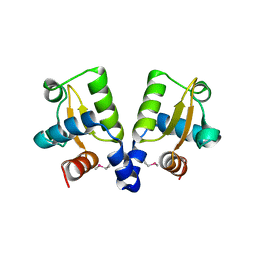 | |
3ULM
 
 | |
3ULO
 
 | |
3V9R
 
 | | Crystal structure of Saccharomyces cerevisiae MHF complex | | 分子名称: | SULFATE ION, Uncharacterized protein YDL160C-A, Uncharacterized protein YOL086W-A | | 著者 | Yang, H, Zhang, T, Zhong, C, Li, H, Zhou, J, Ding, J. | | 登録日 | 2011-12-28 | | 公開日 | 2012-02-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Saccharomyces Cerevisiae MHF Complex Structurally Resembles the Histones (H3-H4)(2) Heterotetramer and Functions as a Heterotetramer
Structure, 20, 2012
|
|
3F1Z
 
 | |
3F5C
 
 | | Structure of Dax-1:LRH-1 complex | | 分子名称: | Nuclear receptor subfamily 0 group B member 1, Nuclear receptor subfamily 5 group A member 2 | | 著者 | Fletterick, R.J, Sablin, E.P. | | 登録日 | 2008-11-03 | | 公開日 | 2008-12-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The structure of corepressor Dax-1 bound to its target nuclear receptor LRH-1.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4IFM
 
 | |
2MWO
 
 | |
6TGT
 
 | |
3CW4
 
 | | Large c-terminal domain of influenza a virus RNA-dependent polymerase PB2 | | 分子名称: | Polymerase basic protein 2 | | 著者 | Kuzuhara, T, Kise, D, Yoshida, H, Horita, T, Murasaki, Y, Utsunomiya, H, Fujiki, H, Tsuge, H. | | 登録日 | 2008-04-21 | | 公開日 | 2009-01-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis of the influenza A virus RNA polymerase PB2 RNA-binding domain containing the pathogenicity-determinant lysine 627 residue
J.Biol.Chem., 284, 2009
|
|
3CZ7
 
 | |
4Z2W
 
 | | Factor Inhibiting HIF in Complex with Fe, and Alpha-Ketoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | 著者 | Taabazuing, C.Y, Garman, S.C, Knapp, M.J. | | 登録日 | 2015-03-30 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Substrate Promotes Productive Gas Binding in the alpha-Ketoglutarate-Dependent Oxygenase FIH.
Biochemistry, 55, 2016
|
|
5D7U
 
 | | Crystal structure of the C-terminal domain of MMTV integrase | | 分子名称: | ISOPROPYL ALCOHOL, Pr160 | | 著者 | Cook, N.J, Pye, V.E, Ballandras-Colas, A, Engelman, A, Cherepanov, P. | | 登録日 | 2015-08-14 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function.
Nature, 530, 2016
|
|
1RW2
 
 | | Three-dimensional structure of Ku80 CTD | | 分子名称: | ATP-dependent DNA helicase II, 80 kDa subunit | | 著者 | Zhang, Z, Hu, W, Cano, L, Lee, T.D, Chen, D.J, Chen, Y. | | 登録日 | 2003-12-15 | | 公開日 | 2003-12-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the C-terminal domain of Ku80 suggests important sites for protein-protein interactions.
STRUCTURE, 12, 2004
|
|
