2Z3C
 
 | |
4K1L
 
 | | 4,4-Dioxo-5,6-dihydro-[1,4,3]oxathiazines, a novel class of 11 beta-HSD1 inhibitors for the treatment of diabetes | | 分子名称: | (4aS,8aR)-N-cyclohexyl-4a,5,6,7,8,8a-hexahydro-4,1,2-benzoxathiazin-3-amine 1,1-dioxide, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Loenze, P, Schimanski-Breves, S, Von der Heyden, C, Engel, C.K. | | 登録日 | 2013-04-05 | | 公開日 | 2014-04-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | 1,1-Dioxo-5,6-dihydro-[4,1,2]oxathiazines, a novel class of 11-HSD1 inhibitors for the treatment of diabetes.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4NZG
 
 | | Crystal Structure of the N-terminal domain of Moloney murine leukemia virus integrase, Northeast Structural Genomics Consortium Target OR3 | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, Integrase p46, ... | | 著者 | Guan, R, Jiang, M, Janjua, H, Maglaqui, M, Zhao, L, Xiao, R, Acton, T.B, Everett, J.K, Roth, M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-12-12 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.152 Å) | | 主引用文献 | X-ray crystal structure of the N-terminal region of Moloney murine leukemia virus integrase and its implications for viral DNA recognition.
Proteins, 85, 2017
|
|
4K00
 
 | | Crystal structure of Slr0204, a 1,4-dihydroxy-2-naphthoyl-CoA thioesterase from Synechocystis | | 分子名称: | 1,2-ETHANEDIOL, 1,4-dihydroxy-2-naphthoyl-CoA hydrolase | | 著者 | Furt, F, Allen, W.J, Widhalm, J.R, Madzelan, P, Rizzo, R.C, Basset, G, Wilson, M.A. | | 登録日 | 2013-04-03 | | 公開日 | 2013-04-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Functional convergence of structurally distinct thioesterases from cyanobacteria and plants involved in phylloquinone biosynthesis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2Z3D
 
 | |
6HAC
 
 | | Crystal structure of [Fe]-hydrogenase (Hmd) holoenzyme from Methanococcus aeolicus (open form) | | 分子名称: | 5,10-methenyltetrahydromethanopterin hydrogenase, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | 著者 | Huang, G, Wagner, T, Wodrich, M.D, Ataka, K, Bill, E, Ermler, U, Hu, X, Shima, S. | | 登録日 | 2018-08-07 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The atomic-resolution crystal structure of activated [Fe]-hydrogenase
Nat Catal, 2019
|
|
1XI0
 
 | | X-ray crystal structure of wild-type Xerocomus chrysenteron lectin XCL | | 分子名称: | lectin | | 著者 | Birck, C, Damian, L, Marty-Detraves, C, Lougarre, A, Schulze-Briese, C, Koehl, P, Fournier, D, Paquereau, L, Samama, J.P. | | 登録日 | 2004-09-21 | | 公開日 | 2005-08-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A new lectin family with structure similarity to actinoporins revealed by the crystal structure of Xerocomus chrysenteron lectin XCL
J.Mol.Biol., 344, 2004
|
|
2Z3E
 
 | |
4O63
 
 | | Co-enzyme Induced Conformational Changes in Bovine Eye Glyceraldehyde 3-Phosphate Dehydrogenase | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Baker, B.Y, Shi, W, Wang, B, Palczewski, K. | | 登録日 | 2013-12-20 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | High-resolution crystal structures of the photoreceptor glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with three and four-bound NAD molecules.
Protein Sci., 23, 2014
|
|
4KGM
 
 | | Bacterial tRNA(HIS) Guanylyltransferase (Thg1)-Like Protein in complex with ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Hyde, S.J, Rao, B.S, Eckenroth, B.E, Jackman, J.E, Doublie, S. | | 登録日 | 2013-04-29 | | 公開日 | 2013-08-14 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.361 Å) | | 主引用文献 | Structural Studies of a Bacterial tRNA(HIS) Guanylyltransferase (Thg1)-Like Protein, with Nucleotide in the Activation and Nucleotidyl Transfer Sites.
Plos One, 8, 2013
|
|
4O9C
 
 | | Crystal structure of Beta-ketothiolase (PhaA) from Ralstonia eutropha H16 | | 分子名称: | Acetyl-CoA acetyltransferase, COENZYME A | | 著者 | Kim, E.J, Kim, J, Kim, S, Kim, K.J. | | 登録日 | 2014-01-02 | | 公開日 | 2014-12-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure and biochemical characterization of PhaA from Ralstonia eutropha, a polyhydroxyalkanoate-producing bacterium.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
3BKD
 
 | | High resolution Crystal structure of Transmembrane domain of M2 protein | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Transmembrane Domain of Matrix protein M2, ... | | 著者 | Stouffer, A.L, Acharya, R, Salom, D. | | 登録日 | 2007-12-06 | | 公開日 | 2008-01-29 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural basis for the function and inhibition of an influenza virus proton channel
Nature, 451, 2008
|
|
4M8H
 
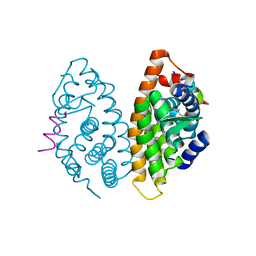 | | CRYSTAL STRUCTURE OF HUMAN RETINOID X RECEPTOR ALPHA-LIGAND BINDING DOMAIN COMPLEX WITH (R)4-METHYL 9cUAB30 AND COACTIVATOR PEPTIDE GRIP-1 | | 分子名称: | (2E,6Z,8E)-3,7-dimethyl-8-[(4R)-4-methyl-3,4-dihydronaphthalen-1(2H)-ylidene]octa-2,6-dienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | 著者 | Xia, G, Smith, C.D, Muccio, D.D. | | 登録日 | 2013-08-13 | | 公開日 | 2014-01-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Methyl-substituted conformationally constrained rexinoid agonists for the retinoid X receptors demonstrate improved efficacy for cancer therapy and prevention.
Bioorg.Med.Chem., 22, 2014
|
|
3A8U
 
 | |
4IMD
 
 | |
4OUB
 
 | | A 2.20 angstroms X-ray crystal structure of E268A 2-aminomucaonate 6-semialdehyde dehydrogenase catalytic intermediate from Pseudomonas fluorescens | | 分子名称: | (2Z,4E)-2-hydroxy-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Huo, L, Liu, A. | | 登録日 | 2014-02-15 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
4IMC
 
 | |
4ZBP
 
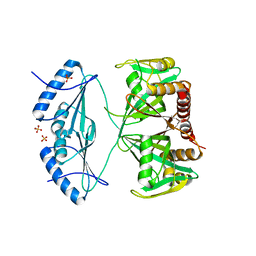 | | Crystal structure of the AMPCPR-bound AtNUDT7 | | 分子名称: | ALPHA-BETA METHYLENE ADP-RIBOSE, Nudix hydrolase 7, SULFATE ION | | 著者 | Tang, Q, Liu, C, Zhong, C, Ding, J. | | 登録日 | 2015-04-15 | | 公開日 | 2015-09-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structures of Arabidopsis thaliana Nudix Hydrolase NUDT7 Reveal a Previously Unobserved Conformation.
Mol Plant, 8, 2015
|
|
7UUK
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with tobramycin | | 分子名称: | Aminocyclitol acetyltransferase ApmA, CHLORIDE ION, TOBRAMYCIN | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2022-04-28 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
3AEK
 
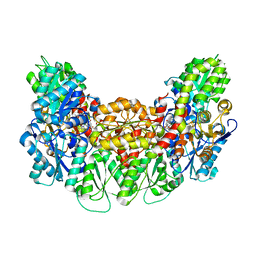 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | 分子名称: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N, ... | | 著者 | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | 登録日 | 2010-02-10 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
4IMF
 
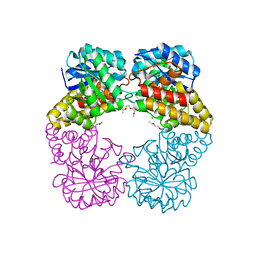 | |
4JTI
 
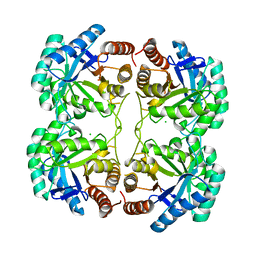 | | Crystal structure of F114R/R117Q/F139G mutant of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | 分子名称: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION | | 著者 | Allison, T.M, Cochrane, F.C, Jameson, G.B, Parker, E.J. | | 登録日 | 2013-03-23 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Examining the Role of Intersubunit Contacts in Catalysis by 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthase.
Biochemistry, 52, 2013
|
|
1BJQ
 
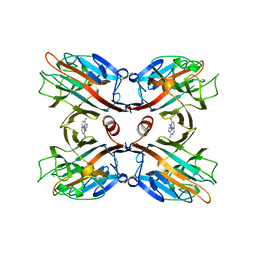 | | THE DOLICHOS BIFLORUS SEED LECTIN IN COMPLEX WITH ADENINE | | 分子名称: | ADENINE, CALCIUM ION, LECTIN, ... | | 著者 | Hamelryck, T.W, Loris, R, Bouckaert, J, Dao-Thi, M.H, Wyns, L, Etzler, M. | | 登録日 | 1998-06-26 | | 公開日 | 1998-12-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Carbohydrate binding, quaternary structure and a novel hydrophobic binding site in two legume lectin oligomers from Dolichos biflorus.
J.Mol.Biol., 286, 1999
|
|
5NX2
 
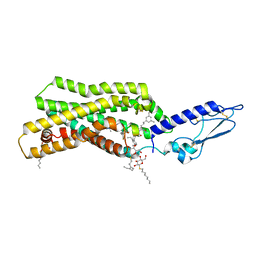 | | Crystal structure of thermostabilised full-length GLP-1R in complex with a truncated peptide agonist at 3.7 A resolution | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucagon-like peptide 1 receptor, ... | | 著者 | Rappas, M, Jazayeri, A, Brown, A.J.H, Kean, J, Errey, J.C, Robertson, N, Fiez-Vandal, C, Andrews, S.P, Congreve, M, Bortolato, A, Mason, J.S, Baig, A.H, Teobald, I, Dore, A.S, Weir, M, Cooke, R.M, Marshall, F.H. | | 登録日 | 2017-05-09 | | 公開日 | 2017-06-14 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Crystal structure of the GLP-1 receptor bound to a peptide agonist.
Nature, 546, 2017
|
|
4LTD
 
 | | Crystal structures of NADH:FMN oxidoreductase (EMOB) - apo form | | 分子名称: | NADH-dependent FMN reductase, PHOSPHATE ION, SULFATE ION | | 著者 | Nissen, M.S, Youn, B, Knowles, B.D, Ballinger, J.W, Jun, S, Belchik, S.M, Xun, L, Kang, C. | | 登録日 | 2013-07-23 | | 公開日 | 2013-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.186 Å) | | 主引用文献 | Crystal structures of NADH:FMN oxidoreductase (EmoB) at different stages of catalysis.
J.Biol.Chem., 283, 2008
|
|
