3ZU2
 
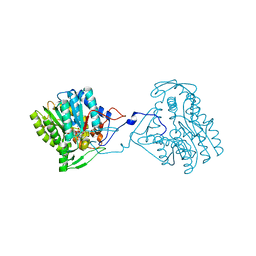 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH (SIRAS) | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, SODIUM ION | | 著者 | Hirschbeck, M.W, Kuper, J, Kisker, C. | | 登録日 | 2011-07-13 | | 公開日 | 2012-01-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
3ZKB
 
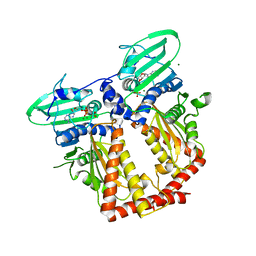 | | CRYSTAL STRUCTURE OF THE ATPASE REGION OF Mycobacterium tuberculosis GyrB WITH AMPPNP | | 分子名称: | DNA GYRASE SUBUNIT B, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Agrawal, A, Roue, M, Spitzfaden, C, Petrella, S, Aubry, A, Volker, C, Mossakowska, D, Hann, M, Bax, B, Mayer, C. | | 登録日 | 2013-01-22 | | 公開日 | 2013-09-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Mycobacterium Tuberculosis DNA Gyrase ATPase Domain Structures Suggest a Dissociative Mechanism that Explains How ATP Hydrolysis is Coupled to Domain Motion.
Biochem.J., 456, 2013
|
|
6E3B
 
 | |
4A0O
 
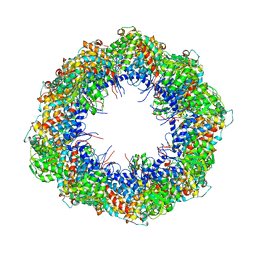 | | Symmetry-free cryo-EM map of TRiC in the nucleotide-free (apo) state | | 分子名称: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | 著者 | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | 登録日 | 2011-09-10 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (10.5 Å) | | 主引用文献 | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
483D
 
 | |
7E80
 
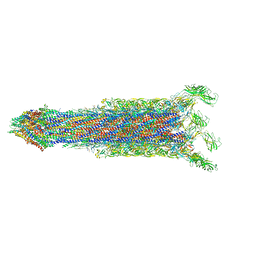 | | Cryo-EM structure of the flagellar rod with hook and export apparatus from Salmonella | | 分子名称: | Flagellar MS ring L1, Flagellar MS ring L2, Flagellar basal body rod protein FlgB, ... | | 著者 | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | 登録日 | 2021-02-28 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.67 Å) | | 主引用文献 | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7E82
 
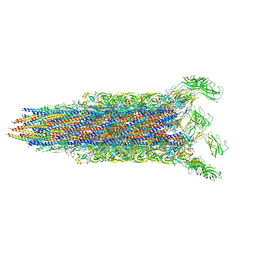 | | Cryo-EM structure of the flagellar rod with partial hook from Salmonella | | 分子名称: | Flagellar MS ring L1, Flagellar MS ring L2, Flagellar basal body rod protein FlgB, ... | | 著者 | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | 登録日 | 2021-02-28 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7EGG
 
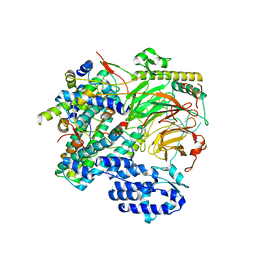 | | TFIID lobe B subcomplex | | 分子名称: | Transcription initiation factor TFIID subunit 10, Transcription initiation factor TFIID subunit 12, Transcription initiation factor TFIID subunit 4, ... | | 著者 | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | 登録日 | 2021-03-24 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
4DQK
 
 | |
7EGF
 
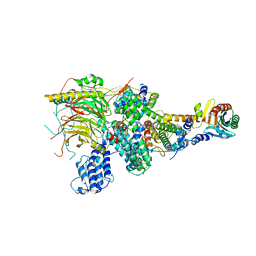 | | TFIID lobe A subcomplex | | 分子名称: | TATA-box-binding protein, Transcription initiation factor TFIID subunit 10, Transcription initiation factor TFIID subunit 11, ... | | 著者 | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | 登録日 | 2021-03-24 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Structural insights into preinitiation complex assembly on core promoters.
Science, 2021
|
|
7EB2
 
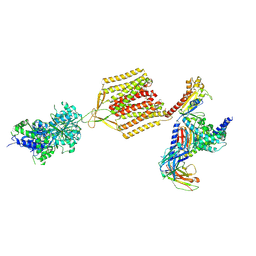 | | Cryo-EM structure of human GABA(B) receptor-Gi protein complex | | 分子名称: | (3S)-5,7-ditert-butyl-3-oxidanyl-3-(trifluoromethyl)-1-benzofuran-2-one, Gamma-aminobutyric acid type B receptor subunit 1, Gamma-aminobutyric acid type B receptor subunit 2, ... | | 著者 | Shen, C, Mao, C, Xu, C, Jin, N, Zhang, H, Shen, D, Shen, Q, Wang, X, Hou, T, Rondard, P, Chen, Z, Pin, J, Zhang, Y, Liu, J. | | 登録日 | 2021-03-08 | | 公開日 | 2021-05-05 | | 最終更新日 | 2021-07-07 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis of GABA B receptor-G i protein coupling.
Nature, 594, 2021
|
|
4DWL
 
 | |
4E0G
 
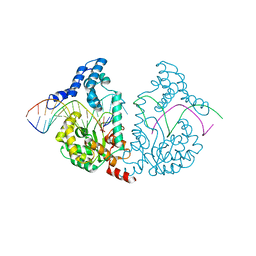 | | Protelomerase tela/DNA hairpin product/vanadate complex | | 分子名称: | DNA (5'-D(*CP*AP*TP*AP*AP*TP*AP*AP*CP*AP*AP*TP*A)-3'), DNA (5'-D(*TP*CP*AP*TP*GP*AP*TP*AP*TP*TP*GP*TP*TP*AP*TP*TP*AP*TP*G)-3'), Protelomerase, ... | | 著者 | Shi, K, Aihara, H. | | 登録日 | 2012-03-03 | | 公開日 | 2013-02-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | An enzyme-catalyzed multistep DNA refolding mechanism in hairpin telomere formation.
Plos Biol., 11, 2013
|
|
7CZV
 
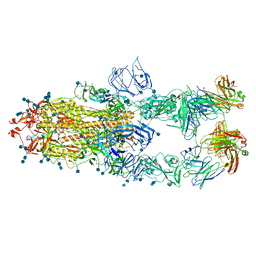 | | S protein of SARS-CoV-2 in complex bound with P5A-1B6_3B | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin heavy variable 3-30-3,chain H of P5A-1B6_3B,Immunoglobulin gamma-1 heavy chain, ... | | 著者 | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | 登録日 | 2020-09-09 | | 公開日 | 2021-03-10 | | 最終更新日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
3XIS
 
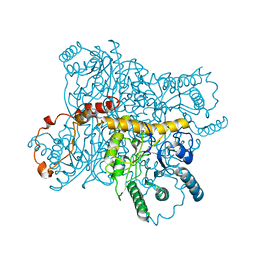 | |
7CZX
 
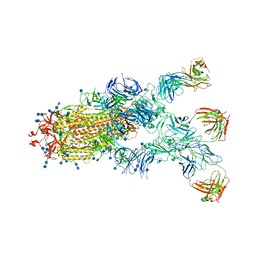 | | S protein of SARS-CoV-2 in complex bound with P5A-1B9 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c168_light_IGKV4-1_IGKJ4,Uncharacterized protein, ... | | 著者 | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | 登録日 | 2020-09-09 | | 公開日 | 2021-03-10 | | 最終更新日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
3ZDS
 
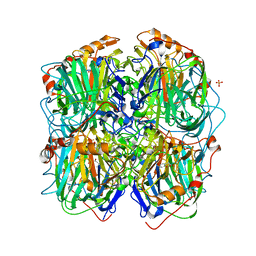 | | Structure of homogentisate 1,2-dioxygenase in complex with reaction intermediates of homogentisate with oxygen. | | 分子名称: | 2-(3,6-DIHYDROXYPHENYL)ACETIC ACID, 2-(6-oxidanyl-3-oxidanylidene-cyclohexa-1,4-dien-1-yl)ethanoic acid, 2-[(6R)-6-(dioxidanyl)-6-oxidanyl-3-oxidanylidene-cyclohexa-1,4-dien-1-yl]ethanoic acid, ... | | 著者 | Jeoung, J.-H, Bommer, M, Lin, T.-Y, Dobbek, H. | | 登録日 | 2012-11-30 | | 公開日 | 2013-07-24 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Visualizing the Substrate-, Superoxo-, Alkylperoxo- and Product-Bound States at the Non-Heme Fe(II) Site of Homogentisate Dioxygenase
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ZLP
 
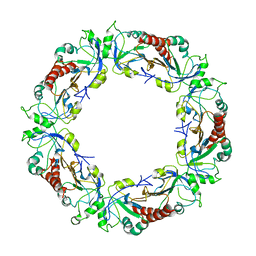 | | Crystal structure of Schistosoma mansoni Peroxiredoxin 1 C48P mutant form with four decamers in the asymmetric unit | | 分子名称: | THIOREDOXIN PEROXIDASE | | 著者 | Saccoccia, F, Angelucci, F, Ardini, M, Boumis, G, Brunori, M, DiLeandro, L, Ippoliti, R, Miele, A.E, Natoli, G, Scotti, S, Bellelli, A. | | 登録日 | 2013-02-04 | | 公開日 | 2013-09-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.515 Å) | | 主引用文献 | Switching between the Alternative Structures and Functions of a 2-Cys Peroxiredoxin, by Site-Directed Mutagenesis
J.Mol.Biol., 425, 2013
|
|
7D98
 
 | |
4DQE
 
 | | Crystal Structure of (G16C/L38C) HIV-1 Protease in Complex with DRV | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, Aspartyl protease | | 著者 | Schiffer, C.A, Mittal, S. | | 登録日 | 2012-02-15 | | 公開日 | 2012-03-07 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Hydrophobic core flexibility modulates enzyme activity in HIV-1 protease.
J.Am.Chem.Soc., 134, 2012
|
|
7D6E
 
 | |
7D6D
 
 | |
8G4Z
 
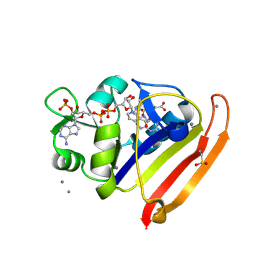 | | E. coli DHFR complex with NADP+ and folate: EF-X off model by Laue diffraction (no electric field) | | 分子名称: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | 著者 | Greisman, J.B, Dalton, K.M, Brookner, D.E, Klureza, M.A, Hekstra, D.R. | | 登録日 | 2023-02-10 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Perturbative diffraction methods resolve a conformational switch that facilitates a two-step enzymatic mechanism.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8G50
 
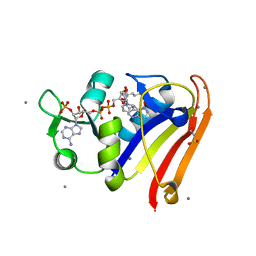 | | E. coli DHFR complex with NADP+ and folate: EF-X excited state model by Laue diffraction (electric field along b axis; 8-fold extrapolation of structure factor differences) | | 分子名称: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | 著者 | Greisman, J.B, Dalton, K.M, Brookner, D.E, Klureza, M.A, Hekstra, D.R. | | 登録日 | 2023-02-10 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Perturbative diffraction methods resolve a conformational switch that facilitates a two-step enzymatic mechanism.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7DKZ
 
 | | Structure of plant photosystem I-light harvesting complex I supercomplex | | 分子名称: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | 著者 | Wang, J, Yu, L.J, Wang, W. | | 登録日 | 2020-11-25 | | 公開日 | 2021-03-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.393 Å) | | 主引用文献 | Structure of plant photosystem I-light harvesting complex I supercomplex at 2.4 angstrom resolution.
J Integr Plant Biol, 63, 2021
|
|
