6R91
 
 | | Cryo-EM structure of NCP_THF2(-3)-UV-DDB | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R8Y
 
 | | Cryo-EM structure of NCP-6-4PP(-1)-UV-DDB | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R93
 
 | | Cryo-EM structure of NCP-6-4PP | | 分子名称: | Histone H2A type 1-B/E, Histone H2B type 1-J, Histone H3.1, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R92
 
 | | Cryo-EM structure of NCP-THF2(+1)-UV-DDB class B | | 分子名称: | DNA damage-binding protein 1,DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R8Z
 
 | | Cryo-EM structure of NCP_THF2(-1)-UV-DDB | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R94
 
 | | Cryo-EM structure of NCP_THF2(-3) | | 分子名称: | Histone H2A type 1-B/E, Histone H2B type 1-J, Histone H3.1, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R90
 
 | | Cryo-EM structure of NCP-THF2(+1)-UV-DDB class A | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
1BQM
 
 | | HIV-1 RT/HBY 097 | | 分子名称: | (S)-4-ISOPROPOXYCARBONYL-6-METHOXY-3-METHYLTHIOMETHYL-3,4-DIHYDROQUINOXALIN-2(1H)-THIONE, REVERSE TRANSCRIPTASE | | 著者 | Hsiou, Y, Das, K, Ding, J, Arnold, E. | | 登録日 | 1998-08-17 | | 公開日 | 1999-01-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structures of Tyr188Leu mutant and wild-type HIV-1 reverse transcriptase complexed with the non-nucleoside inhibitor HBY 097: inhibitor flexibility is a useful design feature for reducing drug resistance.
J.Mol.Biol., 284, 1998
|
|
1BQN
 
 | | TYR 188 LEU HIV-1 RT/HBY 097 | | 分子名称: | (S)-4-ISOPROPOXYCARBONYL-6-METHOXY-3-METHYLTHIOMETHYL-3,4-DIHYDROQUINOXALIN-2(1H)-THIONE, REVERSE TRANSCRIPTASE | | 著者 | Hsiou, Y, Das, K, Ding, J, Arnold, E. | | 登録日 | 1998-08-17 | | 公開日 | 1999-01-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structures of Tyr188Leu mutant and wild-type HIV-1 reverse transcriptase complexed with the non-nucleoside inhibitor HBY 097: inhibitor flexibility is a useful design feature for reducing drug resistance.
J.Mol.Biol., 284, 1998
|
|
2IC3
 
 | |
3V4I
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with DNA and AZTTP | | 分子名称: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*TP*GP*GP*AP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | 著者 | Das, K, Martinez, S.E, Arnold, E. | | 登録日 | 2011-12-15 | | 公開日 | 2012-01-18 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.7983 Å) | | 主引用文献 | HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3V6D
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) cross-linked with AZT-terminated DNA | | 分子名称: | DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*TP*GP*GP*AP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | 著者 | Das, K, Martinez, S.E, Arnold, E. | | 登録日 | 2011-12-19 | | 公開日 | 2012-01-18 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.7048 Å) | | 主引用文献 | HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism.
Nat.Struct.Mol.Biol., 19, 2012
|
|
7T5A
 
 | | Crystal structure of the molybdate-binding periplasmic protein ModA from the bacteria Pseudomonsa aeruginosa in tungstate-bound form | | 分子名称: | AMMONIUM ION, Molybdate-binding periplasmic protein ModA, TUNGSTATE(VI)ION | | 著者 | Ngu, D.H.Y, Luo, Z, Lim, B.Y.J, Kobe, B. | | 登録日 | 2021-12-11 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | The Impact of Chromate on Pseudomonas aeruginosa Molybdenum Homeostasis.
Front Microbiol, 13, 2022
|
|
7T4Z
 
 | | Crystal structure of the molybdate-binding periplasmic protein ModA from the bacteria Pseudomonsa aeruginosa in ligand-free form | | 分子名称: | AMMONIUM ION, GLYCEROL, Molybdate-binding periplasmic protein, ... | | 著者 | Ngu, D.H.Y, Luo, Z, Lim, B.Y.J, Kobe, B. | | 登録日 | 2021-12-10 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | The Impact of Chromate on Pseudomonas aeruginosa Molybdenum Homeostasis.
Front Microbiol, 13, 2022
|
|
7T50
 
 | | Crystal structure of the molybdate-binding periplasmic protein ModA from the bacteria Pseudomonsa aeruginosa in chromate-bound form | | 分子名称: | AMMONIUM ION, Chromate, GLYCEROL, ... | | 著者 | Ngu, D.H.Y, Luo, Z, Lim, B.Y.J, Kobe, B. | | 登録日 | 2021-12-11 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The Impact of Chromate on Pseudomonas aeruginosa Molybdenum Homeostasis.
Front Microbiol, 13, 2022
|
|
7T51
 
 | | Crystal structure of the molybdate-binding periplasmic protein ModA from the bacteria Pseudomonsa aeruginosa in molybdate-bound form | | 分子名称: | AMMONIUM ION, GLYCEROL, MOLYBDATE ION, ... | | 著者 | Ngu, D.H.Y, Luo, Z, Lim, B.Y.J, Kobe, B. | | 登録日 | 2021-12-11 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Impact of Chromate on Pseudomonas aeruginosa Molybdenum Homeostasis.
Front Microbiol, 13, 2022
|
|
7PII
 
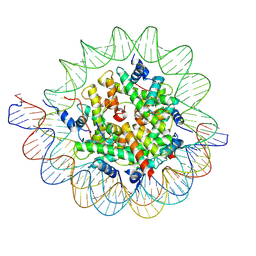 | | Structure of the human CCAN CENP-A alpha-satellite complex | | 分子名称: | Centromere protein C, DNA (122-MER), DNA (123-MER), ... | | 著者 | Yatskevich, S, Muir, K.W, Bellini, D, Barford, D. | | 登録日 | 2021-08-19 | | 公開日 | 2022-05-25 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
2R7J
 
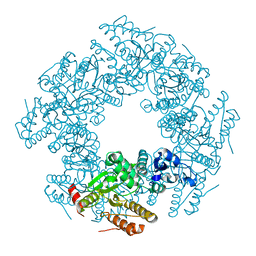 | |
3MKM
 
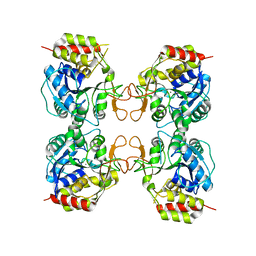 | | Crystal structure of the E. coli pyrimidine nucleoside hydrolase YeiK (Apo-form) | | 分子名称: | CALCIUM ION, Putative uncharacterized protein YeiK | | 著者 | Garau, G, Fornili, A, Giabbai, B, Degano, M. | | 登録日 | 2010-04-15 | | 公開日 | 2010-12-01 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Energy Landscapes Associated with Macromolecular Conformational Changes from Endpoint Structures
J.Am.Chem.Soc., 132, 2010
|
|
2R7P
 
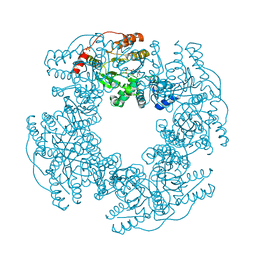 | | Crystal Structure of H225A NSP2 and AMPPNP complex | | 分子名称: | Non-structural RNA-binding protein 35, PHOSPHATE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Kumar, M, Prasad, B.V.V. | | 登録日 | 2007-09-09 | | 公開日 | 2007-10-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystallographic and Biochemical Analysis of Rotavirus NSP2 with Nucleotides Reveals a Nucleoside Diphosphate Kinase-Like Activity
J.Virol., 81, 2007
|
|
5TXL
 
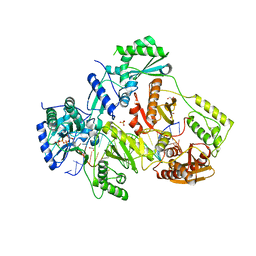 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING DATP | | 分子名称: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG) P*CP*GP*CP*CP*GP)-3'), ... | | 著者 | Das, K, Martinez, S.M, Arnold, E. | | 登録日 | 2016-11-17 | | 公開日 | 2017-04-05 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Antimicrob. Agents Chemother., 61, 2017
|
|
2R7C
 
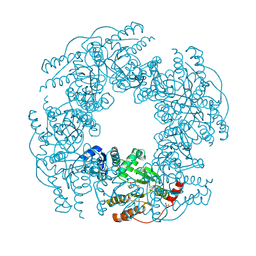 | |
5TXP
 
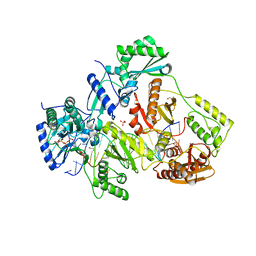 | | STRUCTURE OF Q151M complex (A62V, V75I, F77L, F116Y, Q151M) mutant HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING DDATP | | 分子名称: | 1,2-ETHANEDIOL, 2',3'-dideoxyadenosine triphosphate, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3'), ... | | 著者 | Das, K, Martinez, S.M, Arnold, E. | | 登録日 | 2016-11-17 | | 公開日 | 2017-04-05 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Antimicrob. Agents Chemother., 61, 2017
|
|
5TXN
 
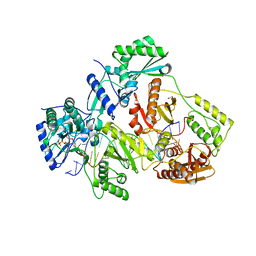 | | STRUCTURE OF Q151M MUTANT HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING DATP | | 分子名称: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3'), ... | | 著者 | Das, K, Martinez, S.M, Arnold, E. | | 登録日 | 2016-11-17 | | 公開日 | 2017-04-05 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Antimicrob. Agents Chemother., 61, 2017
|
|
2R8F
 
 | | Crystal structure of H225A NSP2 and ATP-gS complex | | 分子名称: | Non-structural RNA-binding protein 35, PHOSPHATE ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | 著者 | Kumar, M, Prasad, B.V.V. | | 登録日 | 2007-09-10 | | 公開日 | 2007-10-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystallographic and Biochemical Analysis of Rotavirus NSP2 with Nucleotides Reveals a Nucleoside Diphosphate Kinase-Like Activity
J.Virol., 81, 2007
|
|
