5W0V
 
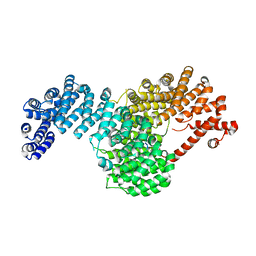 | | Crystal structure of full-length Kluyveromyces lactis Kap123 with histone H4 1-34 | | 分子名称: | Histone H4 1-34, Kap123 | | 著者 | An, S, Yoon, J, Song, J.-J, Cho, U.-S. | | 登録日 | 2017-05-31 | | 公開日 | 2017-11-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.821 Å) | | 主引用文献 | Structure-based nuclear import mechanism of histones H3 and H4 mediated by Kap123.
Elife, 6, 2017
|
|
4OIN
 
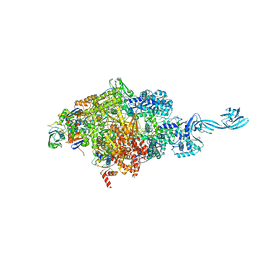 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077 | | 分子名称: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | 著者 | Zhang, Y, Ebright, R.H, Arnold, E. | | 登録日 | 2014-01-20 | | 公開日 | 2014-05-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4OIO
 
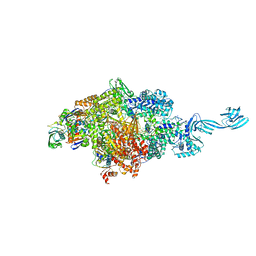 | | Crystal structure of Thermus thermophilus pre-insertion substrate complex for de novo transcription initiation | | 分子名称: | 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ... | | 著者 | Zhang, Y, Ebright, R.H, Arnold, E. | | 登録日 | 2014-01-20 | | 公開日 | 2014-05-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
1FIP
 
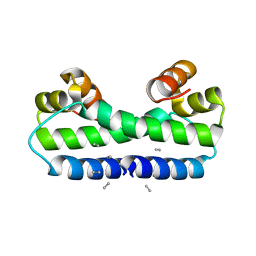 | | THE STRUCTURE OF FIS MUTANT PRO61ALA ILLUSTRATES THAT THE KINK WITHIN THE LONG ALPHA-HELIX IS NOT DUE TO THE PRESENCE OF THE PROLINE RESIDUE | | 分子名称: | FACTOR FOR INVERSION STIMULATION (FIS), UNKNOWN PEPTIDE, POSSIBLY PART OF THE UNOBSERVED RESIDUES IN ENTITY 1 | | 著者 | Yuan, H.S, Wang, S.S, Yang, W.-Z, Finkel, S.E, Johnson, R.C. | | 登録日 | 1994-09-26 | | 公開日 | 1995-02-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The structure of Fis mutant Pro61Ala illustrates that the kink within the long alpha-helix is not due to the presence of the proline residue.
J.Biol.Chem., 269, 1994
|
|
6WMQ
 
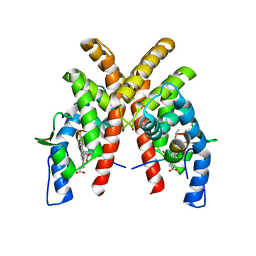 | |
6WMS
 
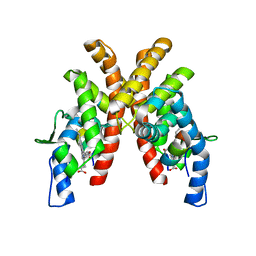 | |
8GAF
 
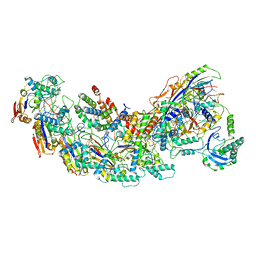 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | 分子名称: | Cas11, Cas5, Cas7, ... | | 著者 | Hu, C, Nam, K.H, Ke, A. | | 登録日 | 2023-02-22 | | 公開日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
4ONM
 
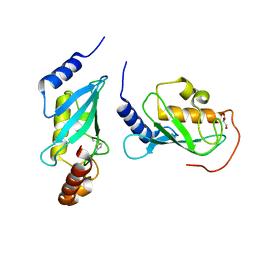 | | Crystal structure of human Mms2/Ubc13 - NSC697923 | | 分子名称: | 2-[(4-methylphenyl)sulfonyl]-5-nitrofuran, GLYCEROL, Ubiquitin-conjugating enzyme E2 N, ... | | 著者 | Hodge, C.D, Edwards, R.A, Glover, J.N.M. | | 登録日 | 2014-01-28 | | 公開日 | 2015-05-06 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Covalent Inhibition of Ubc13 Affects Ubiquitin Signaling and Reveals Active Site Elements Important for Targeting.
Acs Chem.Biol., 10, 2015
|
|
1ETQ
 
 | |
4ONN
 
 | | Crystal structure of human Mms2/Ubc13 - BAY 11-7082 | | 分子名称: | 3-[(4-methylphenyl)sulfonyl]prop-2-enenitrile, GLYCEROL, Ubiquitin-conjugating enzyme E2 N, ... | | 著者 | Hodge, C.D, Edwards, R.A, Glover, J.N.M. | | 登録日 | 2014-01-28 | | 公開日 | 2015-05-06 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Covalent Inhibition of Ubc13 Affects Ubiquitin Signaling and Reveals Active Site Elements Important for Targeting.
Acs Chem.Biol., 10, 2015
|
|
1ETV
 
 | |
1ETW
 
 | |
8HSJ
 
 | | Thermus thermophilus transcription termination factor Rho bound with ADP-BeF3 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | 著者 | Murayama, Y, Ehara, H, Sekine, S. | | 登録日 | 2022-12-19 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis of the transcription termination factor Rho engagement with transcribing RNA polymerase from Thermus thermophilus.
Sci Adv, 9, 2023
|
|
1ETX
 
 | |
1ETK
 
 | |
1ETY
 
 | |
1ETO
 
 | |
2FVQ
 
 | | A Structural Study of the CA Dinucleotide Step in the Integrase Processing Site of Moloney Murine Leukemia Virus | | 分子名称: | 5'-D(*CP*TP*TP*TP*CP*AP*TP*TP*AP*AP*TP*GP*AP*AP*AP*G)-3', reverse transcriptase | | 著者 | Montano, S.P, Cote, M.L, Roth, M.J, Georgiadis, M.M. | | 登録日 | 2006-01-31 | | 公開日 | 2006-12-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of oligonucleotides including the integrase processing site of the Moloney murine leukemia virus.
Nucleic Acids Res., 34, 2006
|
|
8XOT
 
 | |
8XOW
 
 | | Mature virion portal of bacteriophage lambda | | 分子名称: | Head completion protein, Head-tail connector protein FII, Portal protein B, ... | | 著者 | Wang, J.W, Gu, Z.W. | | 登録日 | 2024-01-02 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.32 Å) | | 主引用文献 | Structural morphing in the viral portal vertex of bacteriophage lambda.
J.Virol., 98, 2024
|
|
8XOU
 
 | |
2FVP
 
 | | A Structural Study of the CA Dinucleotide Step in the Integrase Processing Site of Moloney Murine Leukemia Virus | | 分子名称: | 5'-D(*TP*TP*TP*CP*AP*TP*TP*GP*CP*AP*AP*TP*GP*AP*AP*A)-3', Reverse transcriptase | | 著者 | Montano, S.P, Cote, M.L, Roth, M.J, Georgiadis, M.M. | | 登録日 | 2006-01-31 | | 公開日 | 2006-12-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal structures of oligonucleotides including the integrase processing site of the Moloney murine leukemia virus.
Nucleic Acids Res., 34, 2006
|
|
2FVS
 
 | | A Structural Study of the CA Dinucleotide Step in the Integrase Processing Site of Moloney Murine Leukemia Virus | | 分子名称: | 5'-D(*CP*AP*CP*AP*AP*TP*GP*AP*TP*CP*AP*TP*TP*GP*TP*G)-3', reverse transcriptase | | 著者 | Montano, S.P, Cote, M.L, Roth, M.J, Georgiadis, M.M. | | 登録日 | 2006-01-31 | | 公開日 | 2006-12-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structures of oligonucleotides including the integrase processing site of the Moloney murine leukemia virus.
Nucleic Acids Res., 34, 2006
|
|
7Z1L
 
 | | Structure of yeast RNA Polymerase III Pre-Termination Complex (PTC) | | 分子名称: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | 著者 | Girbig, M, Mueller, C.W. | | 登録日 | 2022-02-24 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1N
 
 | | Structure of yeast RNA Polymerase III Delta C53-C37-C11 | | 分子名称: | CHAPSO, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC2, ... | | 著者 | Girbig, M, Mueller, C.W. | | 登録日 | 2022-02-24 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
