6PSK
 
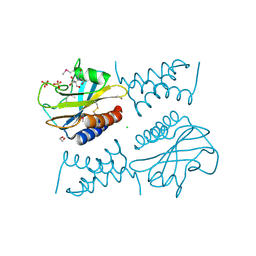 | | Crystal structure of the complex between periplasmic domains of antiholin RI and holin T from T4 phage, in P6522 | | 分子名称: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Antiholin, ... | | 著者 | Kuznetsov, V.B, Krieger, I.V, Sacchettini, J.C. | | 登録日 | 2019-07-12 | | 公開日 | 2020-06-24 | | 最終更新日 | 2020-08-12 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Structural Basis of T4 Phage Lysis Control: DNA as the Signal for Lysis Inhibition.
J.Mol.Biol., 432, 2020
|
|
6HXQ
 
 | |
6FG3
 
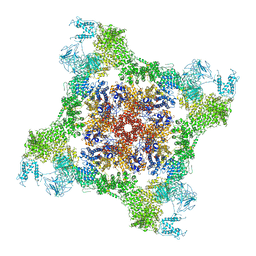 | |
3BCA
 
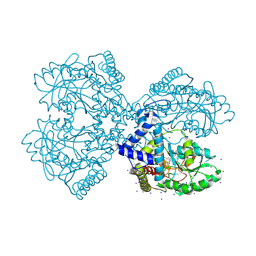 | |
3BIW
 
 | | Crystal structure of the Neuroligin-1/Neurexin-1beta synaptic adhesion complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Arac, D, Boucard, A.A, Ozkan, E, Strop, P, Newell, E, Sudhof, T.C, Brunger, A.T. | | 登録日 | 2007-12-01 | | 公開日 | 2007-12-18 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structures of Neuroligin-1 and the Neuroligin-1/Neurexin-1beta Complex Reveal Specific Protein-Protein and Protein-Ca(2+) Interactions.
Neuron, 56, 2007
|
|
6G3E
 
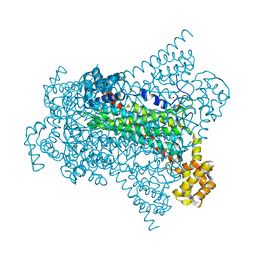 | | Crystal structure of EDDS lyase in complex with formate | | 分子名称: | Argininosuccinate lyase, FORMIC ACID, SODIUM ION | | 著者 | Poddar, H, Thunnissem, A.M.W.H, Poelarends, G.J. | | 登録日 | 2018-03-25 | | 公開日 | 2018-05-16 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Basis for the Catalytic Mechanism of Ethylenediamine- N, N'-disuccinic Acid Lyase, a Carbon-Nitrogen Bond-Forming Enzyme with a Broad Substrate Scope.
Biochemistry, 57, 2018
|
|
6GBQ
 
 | | Crystal Structure of the oligomerization domain of Vp35 from Reston virus | | 分子名称: | Polymerase cofactor VP35 | | 著者 | Zinzula, L, Nagy, I, Orsini, M, Weyher-Stingl, E, Baumeister, W, Bracher, A. | | 登録日 | 2018-04-16 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Structures of Ebola and Reston Virus VP35 Oligomerization Domains and Comparative Biophysical Characterization in All Ebolavirus Species.
Structure, 27, 2019
|
|
6ZVP
 
 | | Atomic model of the EM-based structure of the full-length tyrosine hydroxylase in complex with dopamine (residues 40-497) in which the regulatory domain (residues 40-165) has been included only with the backbone atoms | | 分子名称: | FE (III) ION, L-DOPAMINE, Tyrosine 3-monooxygenase | | 著者 | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | 登録日 | 2020-07-27 | | 公開日 | 2021-11-17 | | 最終更新日 | 2022-02-02 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
6ZZU
 
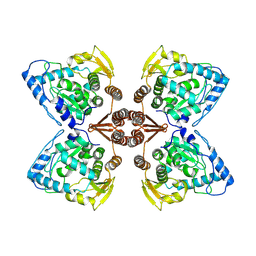 | | Partial structure of the substrate-free tyrosine hydroxylase (apo-TH). | | 分子名称: | FE (III) ION, Tyrosine 3-monooxygenase | | 著者 | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | 登録日 | 2020-08-05 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
7A2G
 
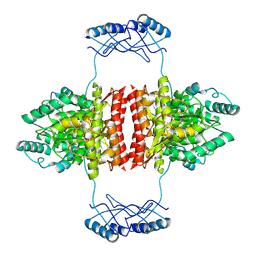 | | Full-length structure of the substrate-free tyrosine hydroxylase (apo-TH). | | 分子名称: | FE (III) ION, Tyrosine 3-monooxygenase | | 著者 | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Flydal, M.I, Martinez, A, Valpuesta, J.M. | | 登録日 | 2020-08-17 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
5L4M
 
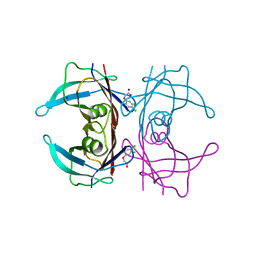 | | Crystal Structure of Human Transthyretin in Complex with 3,5,6-Trichloro-2-pyridinyloxyacetic acid (Triclopyr) | | 分子名称: | SODIUM ION, Transthyretin, Triclopyr | | 著者 | Grundstrom, C, Hall, M, Zhang, J, Olofsson, A, Andersson, P, Sauer-Eriksson, A.E. | | 登録日 | 2016-05-25 | | 公開日 | 2016-10-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.581 Å) | | 主引用文献 | Structure-Based Virtual Screening Protocol for in Silico Identification of Potential Thyroid Disrupting Chemicals Targeting Transthyretin.
Environ. Sci. Technol., 50, 2016
|
|
6NZY
 
 | |
1YAB
 
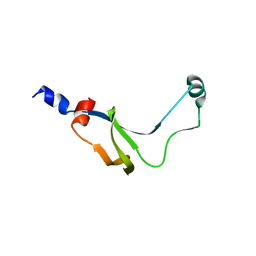 | | Structure of T. maritima FliN flagellar rotor protein | | 分子名称: | chemotaxis protein | | 著者 | Hill, C.P, Blair, D.F, Brown, P.N, Mathews, M.A.A, Joss, L.A. | | 登録日 | 2004-12-17 | | 公開日 | 2005-06-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Crystal Structure of the Flagellar Rotor Protein FliN from Thermotoga maritima
J.BACTERIOL., 187, 2005
|
|
1ROZ
 
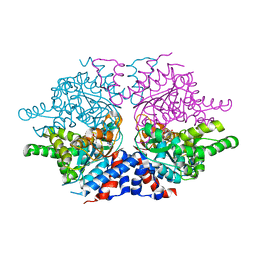 | | Deoxyhypusine synthase holoenzyme in its low ionic strength, high pH crystal form | | 分子名称: | Deoxyhypusine synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Umland, T.C, Wolff, E.C, Park, M.-H, Davies, D.R. | | 登録日 | 2003-12-02 | | 公開日 | 2004-07-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | A New Crystal Structure of Deoxyhypusine Synthase Reveals the Configuration of the Active Enzyme and of an Enzyme-NAD-Inhibitor Ternary Complex
J.Biol.Chem., 279, 2004
|
|
3BCB
 
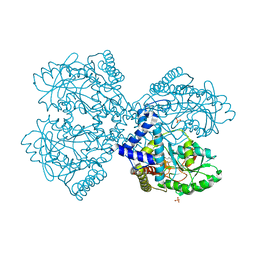 | |
6FOO
 
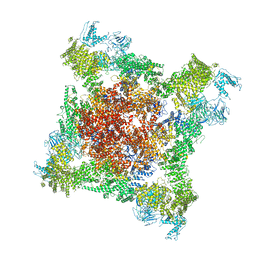 | |
6UCU
 
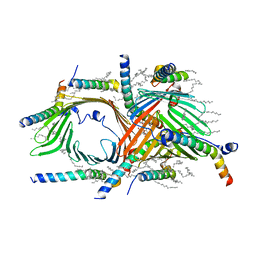 | |
6J6T
 
 | | Crystal Structure of HDA15 HD domain | | 分子名称: | Histone deacetylase 15, POTASSIUM ION, SULFATE ION, ... | | 著者 | Cheng, Y.S, Hsu, J.C, Hung, H.C, Liu, T.C. | | 登録日 | 2019-01-15 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Structure of Arabidopsis HISTONE DEACETYLASE15.
Plant Physiol., 184, 2020
|
|
7B5T
 
 | |
6VME
 
 | | Human ESCRT-I heterotetramer headpiece | | 分子名称: | Multivesicular body subunit 12A, Tumor susceptibility gene 101 protein, Vacuolar protein sorting-associated protein 28 homolog, ... | | 著者 | Flower, T.G, Hurley, J.H, Tjahjono, N. | | 登録日 | 2020-01-27 | | 公開日 | 2020-05-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | A helical assembly of human ESCRT-I scaffolds reverse-topology membrane scission.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8QVD
 
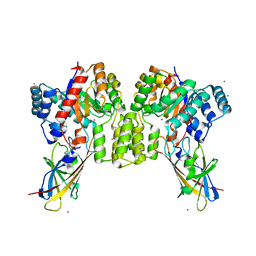 | | Deinococcus aerius TR0125 C-glucosyl deglycosidase (CGD), wild type crystal cryoprotected with glycerol | | 分子名称: | CADMIUM ION, DUF6379 domain-containing protein, Xylose isomerase-like TIM barrel domain-containing protein | | 著者 | Furlanetto, V, Kalyani, D.C, Kostelac, A, Haltrich, D, Hallberg, B.M, Divne, C. | | 登録日 | 2023-10-17 | | 公開日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural and Functional Characterization of a Gene Cluster Responsible for Deglycosylation of C-glucosyl Flavonoids and Xanthonoids by Deinococcus aerius.
J.Mol.Biol., 436, 2024
|
|
1RLZ
 
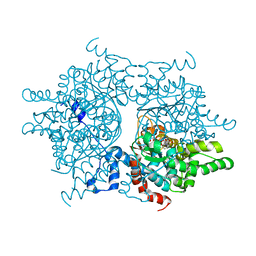 | | Deoxyhypusine synthase holoenzyme in its high ionic strength, low pH crystal form | | 分子名称: | Deoxyhypusine synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Umland, T.C, Wolff, E.C, Park, M.-H, Davies, D.R. | | 登録日 | 2003-11-26 | | 公開日 | 2004-07-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | A New Crystal Structure of Deoxyhypusine Synthase Reveals the Configuration of the Active Enzyme and of an Enzyme-NAD-Inhibitor Ternary Complex
J.Biol.Chem., 279, 2004
|
|
6GBR
 
 | | Crystal Structure of the oligomerization domain of VP35 from Reston virus, mercury derivative | | 分子名称: | MERCURIBENZOIC ACID, Polymerase cofactor VP35 | | 著者 | Zinzula, L, Nagy, I, Orsini, M, Weyher-Stingl, E, Baumeister, W, Bracher, A. | | 登録日 | 2018-04-16 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Structures of Ebola and Reston Virus VP35 Oligomerization Domains and Comparative Biophysical Characterization in All Ebolavirus Species.
Structure, 27, 2019
|
|
2GN1
 
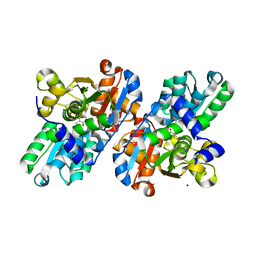 | |
6VWF
 
 | |
