1SCQ
 
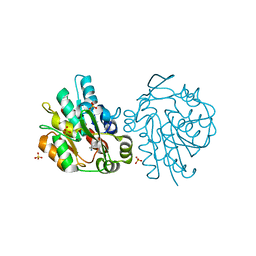 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis in complex with acetonecyanohydrin | | 分子名称: | (S)-acetone-cyanohydrin lyase, 2-HYDROXY-2-METHYLPROPANENITRILE, SULFATE ION | | 著者 | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | 登録日 | 2004-02-12 | | 公開日 | 2004-06-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
1MA9
 
 | | Crystal structure of the complex of human vitamin D binding protein and rabbit muscle actin | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Alpha Skeletal Muscle, ... | | 著者 | Verboven, C, Bogaerts, I, Waelkens, E, Rabijns, A, Van Baelen, H, Bouillon, R, De Ranter, C. | | 登録日 | 2002-08-02 | | 公開日 | 2003-02-04 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Actin-DBP: the perfect structural fit?
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1FO8
 
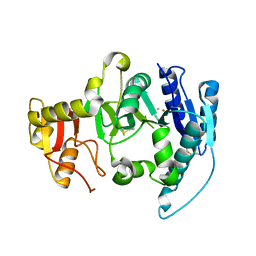 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINYLTRANSFERASE I | | 分子名称: | ALPHA-1,3-MANNOSYL-GLYCOPROTEIN BETA-1,2-N-ACETYLGLUCOSAMINYLTRANSFERASE, METHYL MERCURY ION | | 著者 | Unligil, U.M, Zhou, S, Yuwaraj, S, Sarkar, M, Schachter, H, Rini, J.M. | | 登録日 | 2000-08-26 | | 公開日 | 2001-04-25 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | X-ray crystal structure of rabbit N-acetylglucosaminyltransferase I: catalytic mechanism and a new protein superfamily.
EMBO J., 19, 2000
|
|
3IR2
 
 | |
1SCI
 
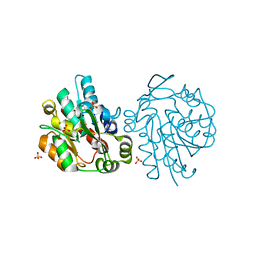 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis | | 分子名称: | (S)-acetone-cyanohydrin lyase, SULFATE ION | | 著者 | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | 登録日 | 2004-02-12 | | 公開日 | 2004-06-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
1SCK
 
 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis in complex with acetone | | 分子名称: | (S)-acetone-cyanohydrin lyase, ACETONE, SULFATE ION | | 著者 | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | 登録日 | 2004-02-12 | | 公開日 | 2004-06-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
4E86
 
 | |
4K0E
 
 | | X-ray crystal structure of a heavy metal efflux pump, crystal form II | | 分子名称: | Heavy metal cation tricomponent efflux pump ZneA(CzcA-like), ZINC ION | | 著者 | Pak, J.E, Ngonlong Ekende, E, Vandenbussche, G, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | 登録日 | 2013-04-03 | | 公開日 | 2013-10-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.713 Å) | | 主引用文献 | Structures of intermediate transport states of ZneA, a Zn(II)/proton antiporter.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4K0J
 
 | | X-ray crystal structure of a heavy metal efflux pump, crystal form I | | 分子名称: | Heavy metal cation tricomponent efflux pump ZneA(CzcA-like), ZINC ION | | 著者 | Pak, J.E, Stroud, R.M, Ngonlong Ekende, E, Vandenbussche, G, Center for Structures of Membrane Proteins (CSMP) | | 登録日 | 2013-04-04 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structures of intermediate transport states of ZneA, a Zn(II)/proton antiporter.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1FX9
 
 | | CARBOXYLIC ESTER HYDROLASE COMPLEX (DIMERIC PLA2 + MJ33 INHIBITOR + SULPHATE IONS) | | 分子名称: | 1-HEXADECYL-3-TRIFLUOROETHYL-SN-GLYCERO-2-PHOSPHATE METHANE, CALCIUM ION, PHOSPHOLIPASE A2, ... | | 著者 | Pan, Y.H, Epstein, T.M, Jain, M.K, Bahnson, B.J. | | 登録日 | 2000-09-25 | | 公開日 | 2001-09-25 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Five coplanar anion binding sites on one face of phospholipase A2: relationship to interface binding.
Biochemistry, 40, 2001
|
|
3QF6
 
 | | Neutron structure of type-III Antifreeze Protein allows the reconstruction of AFP-ice interface | | 分子名称: | Type-3 ice-structuring protein HPLC 12 | | 著者 | Howard, E.I, Blakeley, M.P, Haertlein, M, Petit-Haertlein, I, Mitschler, A, Fisher, S.J, Cousido-Siah, A, Salvay, A.G, Popov, A, Muller-Dieckmann, C, Petrova, T, Podjarny, A. | | 登録日 | 2011-01-21 | | 公開日 | 2011-06-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | NEUTRON DIFFRACTION (1.85 Å) | | 主引用文献 | Neutron structure of type-III antifreeze protein allows the reconstruction of AFP-ice interface.
J.Mol.Recognit., 24, 2011
|
|
1SC9
 
 | | Hydroxynitrile Lyase from Hevea brasiliensis in complex with the natural substrate acetone cyanohydrin | | 分子名称: | (S)-acetone-cyanohydrin lyase, 2-HYDROXY-2-METHYLPROPANENITRILE, SULFATE ION | | 著者 | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | 登録日 | 2004-02-12 | | 公開日 | 2004-06-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
1BNO
 
 | | NMR SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | DNA POLYMERASE BETA | | 著者 | Liu, D.-J, Prasad, R, Wilson, S.H, Derose, E.F, Mullen, G.P. | | 登録日 | 1996-04-25 | | 公開日 | 1996-12-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional solution structure of the N-terminal domain of DNA polymerase beta and mapping of the ssDNA interaction interface.
Biochemistry, 35, 1996
|
|
1BNP
 
 | | NMR SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA, 55 STRUCTURES | | 分子名称: | DNA POLYMERASE BETA | | 著者 | Liu, D.-J, Prasad, R, Wilson, S.H, Derose, E.F, Mullen, G.P. | | 登録日 | 1996-04-25 | | 公開日 | 1996-12-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional solution structure of the N-terminal domain of DNA polymerase beta and mapping of the ssDNA interaction interface.
Biochemistry, 35, 1996
|
|
4E83
 
 | |
3U92
 
 | |
4E82
 
 | |
3U94
 
 | |
3U93
 
 | |
1F2T
 
 | | Crystal Structure of ATP-Free RAD50 ABC-ATPase | | 分子名称: | RAD50 ABC-ATPASE | | 著者 | Hopfner, K.P, Karcher, A, Shin, D.S, Craig, L. | | 登録日 | 2000-05-29 | | 公開日 | 2000-08-02 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily.
Cell(Cambridge,Mass.), 101, 2000
|
|
1F2U
 
 | | Crystal Structure of RAD50 ABC-ATPase | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RAD50 ABC-ATPASE | | 著者 | Hopfner, K.P, Karcher, A, Shin, D.S, Craig, L. | | 登録日 | 2000-05-29 | | 公開日 | 2000-08-02 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily.
Cell(Cambridge,Mass.), 101, 2000
|
|
3N2C
 
 | | Crystal structure of prolidase eah89906 complexed with n-methylphosphonate-l-proline | | 分子名称: | 1-[(R)-hydroxy(methyl)phosphoryl]-L-proline, PROLIDASE, ZINC ION | | 著者 | Patskovsky, Y, Xu, C, Sauder, J.M, Burley, S.K, Raushel, F.M, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-05-17 | | 公開日 | 2010-06-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
3N5C
 
 | |
6NRB
 
 | | hTRiC-hPFD Class2 | | 分子名称: | Prefoldin subunit 1, Prefoldin subunit 2, Prefoldin subunit 3, ... | | 著者 | Gestaut, D.R, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | 登録日 | 2019-01-23 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (8.7 Å) | | 主引用文献 | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
6NRA
 
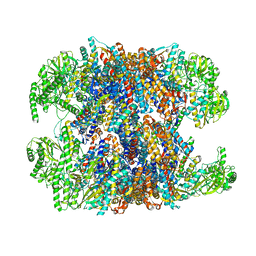 | | hTRiC-hPFD Class1 (No PFD) | | 分子名称: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | 著者 | Gestaut, D.R, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | 登録日 | 2019-01-23 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (7.7 Å) | | 主引用文献 | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
