4N6U
 
 | | Adhiron: a stable and versatile peptide display scaffold - truncated adhiron | | 分子名称: | Adhiron | | 著者 | Mcpherson, M, Tomlinson, D, Owen, R.L, Nettleship, J.E, Owens, R.J. | | 登録日 | 2013-10-14 | | 公開日 | 2014-04-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.251 Å) | | 主引用文献 | Adhiron: a stable and versatile peptide display scaffold for molecular recognition applications.
Protein Eng.Des.Sel., 27, 2014
|
|
4N6T
 
 | | Adhiron: a stable and versatile peptide display scaffold - full length adhiron | | 分子名称: | Adhiron | | 著者 | Mcpherson, M, Tomlinson, D, Owen, R.L, Nettleship, J.E, Owens, R.J. | | 登録日 | 2013-10-14 | | 公開日 | 2014-04-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Adhiron: a stable and versatile peptide display scaffold for molecular recognition applications.
Protein Eng.Des.Sel., 27, 2014
|
|
1RH4
 
 | | RH4 DESIGNED RIGHT-HANDED COILED COIL TETRAMER | | 分子名称: | ISOPROPYL ALCOHOL, RIGHT-HANDED COILED COIL TETRAMER | | 著者 | Harbury, P.B, Plecs, J.J, Tidor, B, Alber, T, Kim, P.S. | | 登録日 | 1998-10-07 | | 公開日 | 1998-12-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | High-resolution protein design with backbone freedom.
Science, 282, 1998
|
|
1SOO
 
 | | ADENYLOSUCCINATE SYNTHETASE INHIBITED BY HYDANTOCIDIN 5'-MONOPHOSPHATE | | 分子名称: | ADENYLOSUCCINATE SYNTHETASE, BETA-MERCAPTOETHANOL, HYDANTOCIDIN-5'-PHOSPHATE, ... | | 著者 | Cowan-Jacob, S.W. | | 登録日 | 1996-05-07 | | 公開日 | 1997-09-04 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The mode of action and the structure of a herbicide in complex with its target: binding of activated hydantocidin to the feedback regulation site of adenylosuccinate synthetase.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1SON
 
 | |
7T03
 
 | | NMR structure of a designed cold unfolding four helix bundle | | 分子名称: | Cold unfolding four helix bundle | | 著者 | Pulavarti, S, Szyperski, T, Yuen, S, Maguire, J, Griffin, J, Kuhlman, B. | | 登録日 | 2021-11-29 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | From Protein Design to the Energy Landscape of a Cold Unfolding Protein.
J.Phys.Chem.B, 126, 2022
|
|
6DKV
 
 | |
6DC1
 
 | | Directed evolutionary changes in Kemp Eliminase KE07 - Crystal 25 round 7 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-nitro-2-oxidanyl-benzenecarbonitrile, ... | | 著者 | Jackson, C.J, Hong, N.-S, Carr, P.D. | | 登録日 | 2018-05-03 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | The evolution of multiple active site configurations in a designed enzyme.
Nat Commun, 9, 2018
|
|
6DNJ
 
 | | Directed evolutionary changes in Kemp Eliminase KE07 - Crystal 28 round 5 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-nitro-1,2-benzoxazole, Kemp eliminase KE07 | | 著者 | Jackson, C.J, Hong, N.-S, Carr, P.D. | | 登録日 | 2018-06-06 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | The evolution of multiple active site configurations in a designed enzyme.
Nat Commun, 9, 2018
|
|
6CT3
 
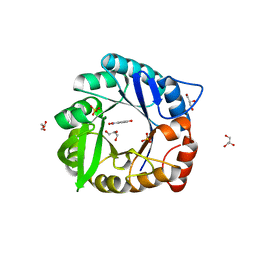 | | Directed evolutionary changes in Kemp Eliminase KE07 - Crystal 27 round 7-2 | | 分子名称: | 5-nitro-2-oxidanyl-benzenecarbonitrile, GLYCEROL, Kemp eliminase, ... | | 著者 | Jackson, C.J, Hong, N.-S, Carr, P.D. | | 登録日 | 2018-03-22 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The evolution of multiple active site configurations in a designed enzyme.
Nat Commun, 9, 2018
|
|
3TES
 
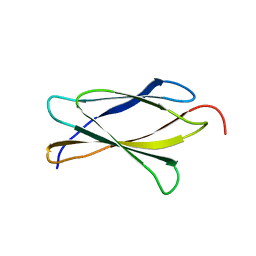 | | Crystal Structure of Tencon | | 分子名称: | Tencon | | 著者 | Luo, J, Jacobs, S, Teplyakov, A, Obmolova, G, O'Neil, K, Gilliland, G. | | 登録日 | 2011-08-15 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Design of novel FN3 domains with high stability by a consensus sequence approach.
Protein Eng.Des.Sel., 25, 2012
|
|
3TEU
 
 | | Crystal structure of fibcon | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, Fibcon | | 著者 | Luo, J, Jacobs, S, Teplyakov, A, Obmolova, G, O'Neil, K, Gilliland, G. | | 登録日 | 2011-08-15 | | 公開日 | 2012-05-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.002 Å) | | 主引用文献 | Design of novel FN3 domains with high stability by a consensus sequence approach.
Protein Eng.Des.Sel., 25, 2012
|
|
6Z1L
 
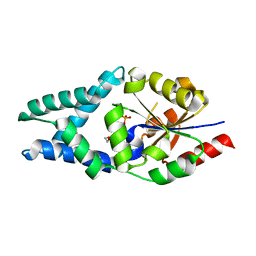 | |
6Z1K
 
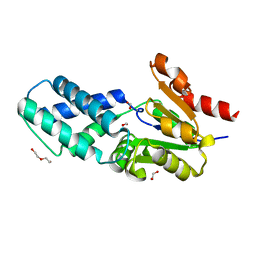 | |
8R3D
 
 | |
8R3B
 
 | |
8R3C
 
 | |
1CLI
 
 | | X-RAY CRYSTAL STRUCTURE OF AMINOIMIDAZOLE RIBONUCLEOTIDE SYNTHETASE (PURM), FROM THE E. COLI PURINE BIOSYNTHETIC PATHWAY, AT 2.5 A RESOLUTION | | 分子名称: | PROTEIN (PHOSPHORIBOSYL-AMINOIMIDAZOLE SYNTHETASE), SULFATE ION | | 著者 | Li, C, Kappock, T.J, Stubbe, J, Weaver, T.M, Ealick, S.E. | | 登録日 | 1999-04-28 | | 公開日 | 1999-10-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | X-ray crystal structure of aminoimidazole ribonucleotide synthetase (PurM), from the Escherichia coli purine biosynthetic pathway at 2.5 A resolution.
Structure Fold.Des., 7, 1999
|
|
1DNL
 
 | | X-RAY STRUCTURE OF ESCHERICHIA COLI PYRIDOXINE 5'-PHOSPHATE OXIDASE COMPLEXED WITH FMN AT 1.8 ANGSTROM RESOLUTION | | 分子名称: | FLAVIN MONONUCLEOTIDE, PHOSPHATE ION, PYRIDOXINE 5'-PHOSPHATE OXIDASE | | 著者 | Safo, M.K, Mathews, I, Musayev, F.N, di Salvo, M.L, Thiel, D.J, Abraham, D.J, Schirch, V. | | 登録日 | 1999-12-16 | | 公開日 | 2000-01-05 | | 最終更新日 | 2017-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | X-ray structure of Escherichia coli pyridoxine 5'-phosphate oxidase complexed with FMN at 1.8 A resolution.
Structure Fold.Des., 8, 2000
|
|
4OFC
 
 | | 2.0 Angstroms X-ray crystal structure of human 2-amino-3-carboxymuconate-6-semialdehye decarboxylase | | 分子名称: | 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase, ZINC ION | | 著者 | Huo, L, Liu, F, Iwaki, H, Chen, L, Hasegawa, Y, Liu, A. | | 登録日 | 2014-01-14 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Human alpha-amino-beta-carboxymuconate-epsilon-semialdehyde decarboxylase (ACMSD): A structural and mechanistic unveiling.
Proteins, 83, 2015
|
|
3U13
 
 | | Crystal Structure of de Novo design of cystein esterase ECH13 at the resolution 1.6A, Northeast Structural Genomics Consortium Target OR51 | | 分子名称: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Kohan, E, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2011-09-29 | | 公開日 | 2011-11-23 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3TWE
 
 | | Crystal Structure of the de novo designed peptide alpha4H | | 分子名称: | ACETYL GROUP, TRIETHYLENE GLYCOL, alpha4H | | 著者 | Buer, B.C, Meagher, J.L, Stuckey, J.A, Marsh, E.N.G. | | 登録日 | 2011-09-21 | | 公開日 | 2012-03-14 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Structural basis for the enhanced stability of highly fluorinated proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6PNR
 
 | | A GH31 family sulfoquinovosidase from E. rectale in complex with aza-sugar inhibitor IFGSQ | | 分子名称: | Alpha-glucosidase, SULFATE ION, [(3~{S},4~{R},5~{R})-4,5-bis(oxidanyl)piperidin-3-yl]methanesulfonic acid | | 著者 | Jarva, M.A, Lingford, J.P, John, A, Goddard-Borger, E.D. | | 登録日 | 2019-07-03 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A GH31 family sulfoquinovosidase from E. rectale in complex with aza-sugar inhibitor IFGSQ
To Be Published
|
|
2CXG
 
 | | CYCLODEXTRIN GLYCOSYLTRANSFERASE COMPLEXED TO THE INHIBITOR ACARBOSE | | 分子名称: | 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | 著者 | Strokopytov, B.V, Uitdehaag, J.C.M, Ruiterkamp, R, Dijkstra, B.W. | | 登録日 | 1998-05-08 | | 公開日 | 1998-10-14 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | X-ray structure of cyclodextrin glycosyltransferase complexed with acarbose. Implications for the catalytic mechanism of glycosidases.
Biochemistry, 34, 1995
|
|
7SGE
 
 | |
