3CCV
 
 | |
3CC7
 
 | |
7L7B
 
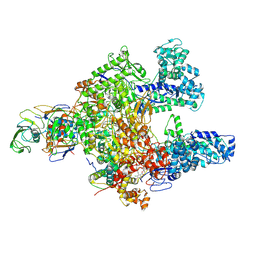 | | Clostridioides difficile RNAP with fidaxomicin | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Boyaci, H, Campbell, E.A, Darst, S.A, Chen, J. | | 登録日 | 2020-12-28 | | 公開日 | 2022-02-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Basis of narrow-spectrum activity of fidaxomicin on Clostridioides difficile.
Nature, 604, 2022
|
|
4NBQ
 
 | | Structure of the polynucleotide phosphorylase (CBU_0852) from Coxiella burnetii | | 分子名称: | Polyribonucleotide nucleotidyltransferase, SULFATE ION | | 著者 | Rudolph, M.J, Cheung, J, Franklin, M.C, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | 登録日 | 2013-10-23 | | 公開日 | 2015-06-17 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.9138 Å) | | 主引用文献 | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3CCE
 
 | |
3C9V
 
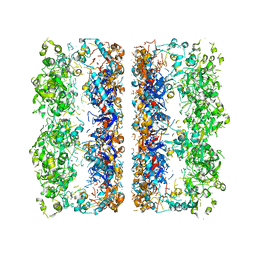 | | C7 Symmetrized Structure of Unliganded GroEL at 4.7 Angstrom Resolution from CryoEM | | 分子名称: | 60 kDa chaperonin | | 著者 | Ludtke, S.J, Baker, M.L, Chen, D.H, Song, J.L, Chuang, D, Chiu, W. | | 登録日 | 2008-02-18 | | 公開日 | 2008-09-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | De Novo Backbone Trace of GroEL from Single Particle Electron Cryomicroscopy.
Structure, 16, 2008
|
|
3CBD
 
 | |
3CC4
 
 | |
7MHF
 
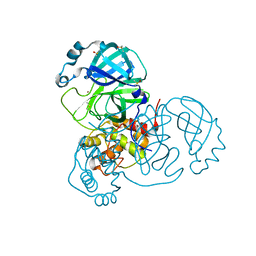 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 100 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
3DV1
 
 | | Crystal structure of human beta-secretase in complex with NVP-ARV999 | | 分子名称: | (2R,4S)-N-butyl-4-[(2S,5S,7R)-2,7-dimethyl-3,15-dioxo-1,4-diazacyclopentadecan-5-yl]-4-hydroxy-2-methylbutanamide, Beta-secretase 1 | | 著者 | Rondeau, J.-M. | | 登録日 | 2008-07-18 | | 公開日 | 2009-02-24 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Macrocyclic peptidomimetic beta-secretase (BACE-1) inhibitors with activity in vivo.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
7MHN
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 277 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1908 Å) | | 主引用文献 | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
4N5Z
 
 | | Crystal structure of aerosol transmissible influenza H5 hemagglutinin mutant (N158D, N224K, Q226L and T318I) from the influenza virus A/Viet Nam/1203/2004 (H5N1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | 著者 | Zhu, X, Wilson, I.A. | | 登録日 | 2013-10-10 | | 公開日 | 2013-11-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.9537 Å) | | 主引用文献 | Hemagglutinin Receptor Specificity and Structural Analyses of Respiratory Droplet-Transmissible H5N1 Viruses.
J.Virol., 88, 2014
|
|
5HDN
 
 | |
7MHL
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 100 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHH
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 277 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1908 Å) | | 主引用文献 | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
2WP1
 
 | | Structure of Brdt bromodomain 2 bound to an acetylated histone H3 peptide | | 分子名称: | BROMODOMAIN TESTIS-SPECIFIC PROTEIN, HISTONE H3 | | 著者 | Moriniere, J, Rousseaux, S, Steuerwald, U, Soler-Lopez, M, Curtet, S, Vitte, A.-L, Govin, J, Gaucher, J, Sadoul, K, Hart, D.J, Krijgsveld, J, Khochbin, S, Mueller, C.W, Petosa, C. | | 登録日 | 2009-08-02 | | 公開日 | 2009-09-22 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Cooperative Binding of Two Acetylation Marks on a Histone Tail by a Single Bromodomain.
Nature, 461, 2009
|
|
5HGL
 
 | | Hexameric HIV-1 CA, open conformation | | 分子名称: | CHLORIDE ION, Capsid protein P24, N-METHYL-NALPHA-[(2-METHYL-1H-INDOL-3-YL)ACETYL]-N-PHENYL-L-PHENYLALANINAMIDE | | 著者 | Price, A.J, Jacques, D.A, James, L.C. | | 登録日 | 2016-01-08 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis.
Nature, 536, 2016
|
|
2X3Y
 
 | | Crystal structure of GmhA from Burkholderia pseudomallei | | 分子名称: | PHOSPHOHEPTOSE ISOMERASE, ZINC ION | | 著者 | Harmer, N.J. | | 登録日 | 2010-01-28 | | 公開日 | 2010-05-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The Structure of Sedoheptulose-7-Phosphate Isomerase from Burkholderia Pseudomallei Reveals a Zinc Binding Site at the Heart of the Active Site.
J.Mol.Biol., 400, 2010
|
|
1QC6
 
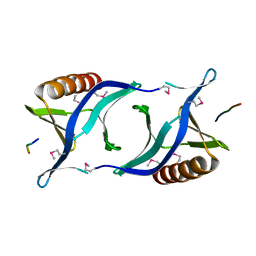 | | EVH1 domain from ENA/VASP-like protein in complex with ACTA peptide | | 分子名称: | EVH1 DOMAIN FROM ENA/VASP-LIKE PROTEIN, PHE-GLU-PHE-PRO-PRO-PRO-PRO-THR-ASP-GLU-GLU | | 著者 | Fedorov, A.A, Fedorov, E.V, Gertler, F.B, Almo, S.C. | | 登録日 | 1999-05-17 | | 公開日 | 1999-05-25 | | 最終更新日 | 2018-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of EVH1, a novel proline-rich ligand-binding module involved in cytoskeletal dynamics and neural function
Nat.Struct.Biol., 6, 1999
|
|
5GL1
 
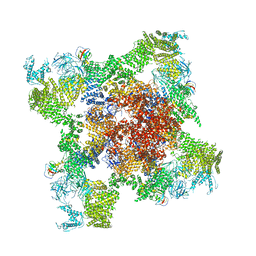 | | Structure of RyR1 in an open state | | 分子名称: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ZINC ION | | 著者 | Bai, X.C, Yan, Z, Wu, J.P, Yan, N. | | 登録日 | 2016-07-07 | | 公開日 | 2016-08-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (5.7 Å) | | 主引用文献 | The Central domain of RyR1 is the transducer for long-range allosteric gating of channel opening
Cell Res., 26, 2016
|
|
5GM2
 
 | | Crystal structure of methyltransferase TleD complexed with SAH and teleocidin A1 | | 分子名称: | (2S,5S)-9-[(3R)-3,7-dimethylocta-1,6-dien-3-yl]-5-(hydroxymethyl)-1-methyl-2-(propan-2-yl)-1,2,4,5,6,8-hexahydro-3H-[1,4]diazonino[7,6,5-cd]indol-3-one, O-methylransferase, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Yu, F, Li, M.J, Xu, C.Y, Zhou, H, Sun, B, Wang, Z.J, Xu, Q, Xie, M.Y, Zuo, G, Huang, P, Wang, Q.S, He, J.H. | | 登録日 | 2016-07-12 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure and enantioselectivity of terpene cyclization in SAM-dependent methyltransferase TleD
Biochem.J., 473, 2016
|
|
4NNU
 
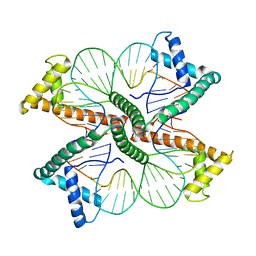 | | Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation | | 分子名称: | DNA1, DNA2, Transcription factor A, ... | | 著者 | Ngo, H.B, Lovely, G.A, Phillips, R, Chan, D.C. | | 登録日 | 2013-11-19 | | 公開日 | 2014-01-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation.
Nat Commun, 5, 2014
|
|
5GAP
 
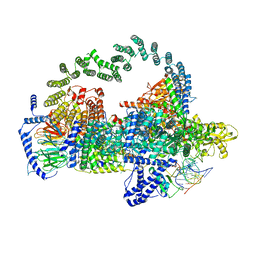 | | Body region of the U4/U6.U5 tri-snRNP | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, Pre-mRNA-processing factor 31, Pre-mRNA-splicing factor 6, ... | | 著者 | Nguyen, T.H.D, Galej, W.P, Oubridge, C, Bai, X.C, Newman, A, Scheres, S, Nagai, K. | | 登録日 | 2015-12-15 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 angstrom resolution.
Nature, 530, 2016
|
|
1OAV
 
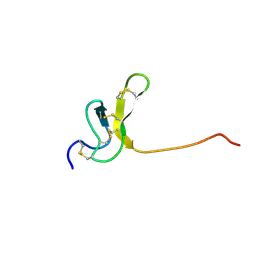 | | OMEGA-AGATOXIN IVA | | 分子名称: | OMEGA-AGATOXIN IVA | | 著者 | Kim, J.I, Konishi, S, Iwai, H, Kohno, T, Gouda, H, Shimada, I, Sato, K, Arata, Y. | | 登録日 | 1995-06-28 | | 公開日 | 1995-10-15 | | 最終更新日 | 2017-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional solution structure of the calcium channel antagonist omega-agatoxin IVA: consensus molecular folding of calcium channel blockers.
J.Mol.Biol., 250, 1995
|
|
3CCM
 
 | |
