5U2T
 
 | |
3O1C
 
 | | High resolution crystal structure of histidine triad nucleotide-binding protein 1 (Hint1) C38A mutant from rabbit complexed with Adenosine | | 分子名称: | ADENOSINE, Histidine triad nucleotide-binding protein 1, SODIUM ION | | 著者 | Dolot, R.M, Ozga, M, Krakowiak, A.K, Nawrot, B, Stec, W.J. | | 登録日 | 2010-07-21 | | 公開日 | 2010-08-04 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | Histidine Triad Nucleotide-binding Protein 1 (HINT-1) Phosphoramidase Transforms Nucleoside 5'-O-Phosphorothioates to Nucleoside 5'-O-Phosphates.
J.Biol.Chem., 285, 2010
|
|
3O1X
 
 | | High resolution crystal structure of histidine triad nucleotide-binding protein 1 (Hint1) C84A mutant from rabbit complexed with adenosine | | 分子名称: | ADENOSINE, Histidine triad nucleotide-binding protein 1, SODIUM ION | | 著者 | Dolot, R.M, Ozga, M, Krakowiak, A.K, Nawrot, B, Stec, W.J. | | 登録日 | 2010-07-22 | | 公開日 | 2010-08-04 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | Histidine Triad Nucleotide-binding Protein 1 (HINT-1) Phosphoramidase Transforms Nucleoside 5'-O-Phosphorothioates to Nucleoside 5'-O-Phosphates.
J.Biol.Chem., 285, 2010
|
|
3O1Z
 
 | | High resolution crystal structure of histidine triad nucleotide-binding protein 1 (Hint1) double cysteine mutant from rabbit | | 分子名称: | Histidine triad nucleotide-binding protein 1 | | 著者 | Dolot, R.M, Ozga, M, Krakowiak, A.K, Nawrot, B, Stec, W.J. | | 登録日 | 2010-07-22 | | 公開日 | 2010-08-04 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Histidine Triad Nucleotide-binding Protein 1 (HINT-1) Phosphoramidase Transforms Nucleoside 5'-O-Phosphorothioates to Nucleoside 5'-O-Phosphates.
J.Biol.Chem., 285, 2010
|
|
4WBF
 
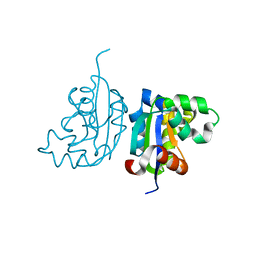 | | Acinetobacter baumannii SDF NDK | | 分子名称: | Nucleoside diphosphate kinase | | 著者 | Hu, Y, Liu, Y. | | 登録日 | 2014-09-03 | | 公開日 | 2015-06-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | Structural and Functional Characterization of Acinetobacter baumannii Nucleoside Diphosphate Kinase
Prog.Biochem.Biophys., 42, 2015
|
|
8TZ4
 
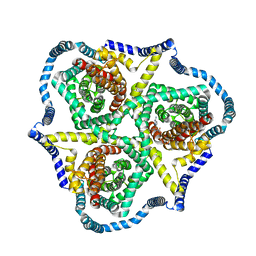 | | Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with GS-441524, subset reconstruction | | 分子名称: | (2~{R},3~{R},4~{S},5~{R})-2-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolane-2-carbonitrile, Sodium/nucleoside cotransporter | | 著者 | Wright, N.J, Lee, S.-Y. | | 登録日 | 2023-08-26 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-09-18 | | 実験手法 | ELECTRON MICROSCOPY (3.23 Å) | | 主引用文献 | Antiviral drug recognition and elevator-type transport motions of CNT3.
Nat.Chem.Biol., 20, 2024
|
|
8TZ2
 
 | |
8TZ7
 
 | |
8TZ1
 
 | | Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with Ribavirin | | 分子名称: | 1-(beta-D-ribofuranosyl)-1H-1,2,4-triazole-3-carboxamide, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, SODIUM ION, ... | | 著者 | Wright, N.J, Lee, S.-Y. | | 登録日 | 2023-08-26 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-09-18 | | 実験手法 | ELECTRON MICROSCOPY (2.54 Å) | | 主引用文献 | Antiviral drug recognition and elevator-type transport motions of CNT3.
Nat.Chem.Biol., 20, 2024
|
|
8TZ5
 
 | |
8TZ6
 
 | |
8TZA
 
 | |
8TZ3
 
 | |
8TZ9
 
 | |
8TZ8
 
 | |
7O62
 
 | |
8TZD
 
 | |
4BY5
 
 | |
4BY4
 
 | |
4X08
 
 | | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms. | | 分子名称: | Eosinophil cationic protein, SULFATE ION | | 著者 | Blanco, J.A, Garcia, J.M, Salazar, V.A, Sanchez, D, Moussauoi, M, Boix, E. | | 登録日 | 2014-11-21 | | 公開日 | 2015-10-07 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms.
To Be Published
|
|
6R4S
 
 | |
6R4T
 
 | |
6RB4
 
 | |
6R5E
 
 | |
1HLW
 
 | | STRUCTURE OF THE H122A MUTANT OF THE NUCLEOSIDE DIPHOSPHATE KINASE | | 分子名称: | NUCLEOSIDE DIPHOSPHATE KINASE | | 著者 | Admiraal, S.J, Meyer, P, Schneider, B, Deville-Bonne, D, Janin, J, Herschlag, D. | | 登録日 | 2000-12-04 | | 公開日 | 2001-02-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Chemical rescue of phosphoryl transfer in a cavity mutant: a cautionary tale for site-directed mutagenesis.
Biochemistry, 40, 2001
|
|
