3UFL
 
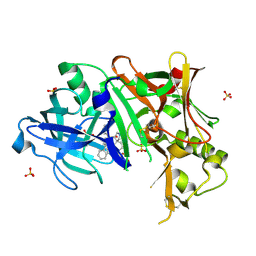 | | Discovery of Pyrrolidine-based b-Secretase Inhibitors: Lead Advancement through Conformational Design for Maintenance of Ligand Binding Efficiency | | 分子名称: | (1R,4'S)-3,4-dihydro-2H-spiro[naphthalene-1,3'-pyrrolidin]-4'-yl[(2S,4R)-2,4-diphenylpiperidin-1-yl]methanone, Beta-secretase 1, GLYCEROL, ... | | 著者 | Allison, T, Munshi, S, Soisson, S.M. | | 登録日 | 2011-11-01 | | 公開日 | 2012-01-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery of pyrrolidine-based beta-secretase inhibitors: Lead advancement through conformational design for maintenance of ligand binding efficiency.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6ZD4
 
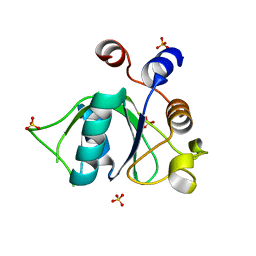 | |
8GH6
 
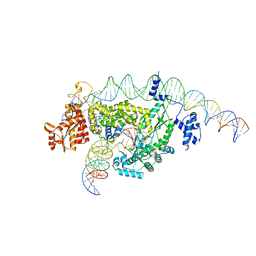 | |
6ZCM
 
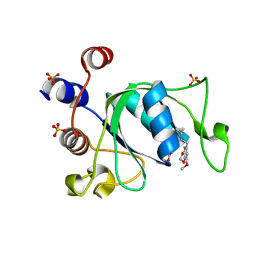 | | Crystal structure of YTHDC1 with compound DHU_DC1_180 | | 分子名称: | 6-[[cyclopropyl-[(7-methoxy-1,3-benzodioxol-5-yl)methyl]amino]methyl]-1~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | 著者 | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | 登録日 | 2020-06-11 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6ZD3
 
 | | Crystal structure of YTHDC1 M438A mutant | | 分子名称: | DI(HYDROXYETHYL)ETHER, SULFATE ION, YTH domain containing 1 | | 著者 | Bedi, R.K, Li, Y, Caflisch, A. | | 登録日 | 2020-06-13 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6Z18
 
 | | Crystal structure of RNA-10mer: CCGG(N4,N4-dimethyl-C)GCCGG; R32 form | | 分子名称: | RNA-10mer: CCGG(N4,N4-dimethyl-C)GCCGG | | 著者 | Ruszkowski, M, Sekula, B, Mao, S, Haruehanroengra, P, Sheng, J. | | 登録日 | 2020-05-12 | | 公開日 | 2020-09-02 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Base pairing, structural and functional insights into N4-methylcytidine (m4C) and N4,N4-dimethylcytidine (m42C) modified RNA.
Nucleic Acids Res., 48, 2020
|
|
6ZCN
 
 | | Crystal structure of YTHDC1 with m6A | | 分子名称: | N6-METHYLADENOSINE-5'-MONOPHOSPHATE, SULFATE ION, YTHDC1 | | 著者 | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | 登録日 | 2020-06-11 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
7RAA
 
 | | Designed StabIL-2 seq15 | | 分子名称: | Interleukin-2, MAGNESIUM ION | | 著者 | Jude, K.M, Chu, A.E, Huang, P.-S, Garcia, K.C. | | 登録日 | 2021-06-30 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Interleukin-2 superkines by computational design.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RA9
 
 | | Designed StabIL-2 seq1 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Interleukin-2, PHOSPHATE ION | | 著者 | Jude, K.M, Chu, A.E, Huang, P.-S, Garcia, K.C. | | 登録日 | 2021-06-30 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Interleukin-2 superkines by computational design.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7R79
 
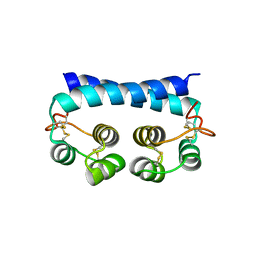 | | Histoplasma capsulatum H88 Calcium Binding Protein 1 (Cbp1) | | 分子名称: | Calcium-binding protein | | 著者 | Herrera, N, Azimova, D, Sil, A, Rosenberg, O.S. | | 登録日 | 2021-06-24 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Cbp1, a fungal virulence factor under positive selection, forms an effector complex that drives macrophage lysis.
Plos Pathog., 18, 2022
|
|
7R6U
 
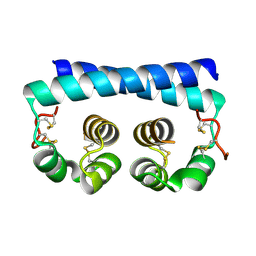 | | Paracoccidioides americana Pb03 Calcium Binding Protein 1 (Cbp1) | | 分子名称: | CBP domain-containing protein | | 著者 | Herrera, N, Azimova, D, Sil, A, Rosenberg, O.S. | | 登録日 | 2021-06-23 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Cbp1, a fungal virulence factor under positive selection, forms an effector complex that drives macrophage lysis.
Plos Pathog., 18, 2022
|
|
7RDW
 
 | |
6Z1H
 
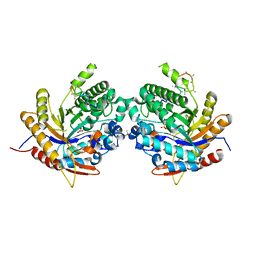 | | Ancestral glycosidase (family 1) | | 分子名称: | ANCESTRAL RECONSTRUCTED GLYCOSIDASE, GLYCEROL, ISOPROPYL ALCOHOL, ... | | 著者 | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M, Gamiz-Arco, G, Gutierrez-Rus, L, Ibarra-Molero, B, Hoshino, Y, Petrovic, D, Romero-Rivera, A, Seelig, B, Kamerlin, S.C.L, Gaucher, E.A. | | 登録日 | 2020-05-13 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Heme-binding enables allosteric modulation in an ancient TIM-barrel glycosidase.
Nat Commun, 12, 2021
|
|
6ZD5
 
 | |
6ZDA
 
 | |
6ZD8
 
 | |
8G4S
 
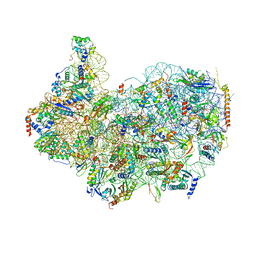 | | 40S ribosomal subunit of the 80S Giardia intestinalis assemblage A ribosome with Emetine bound in V2 conformation with mRNA and three tRNAs. | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S25, ... | | 著者 | Eiler, D.R, Wimberly, B.T, Bilodeau, D.Y, Rissland, O.S, Kieft, J.S. | | 登録日 | 2023-02-10 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | The Giardia lamblia ribosome structure reveals divergence in several biological pathways and the mode of emetine function.
Structure, 32, 2024
|
|
7R0X
 
 | |
6Z6Q
 
 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH) oxygenase and TPR domains in complex with manganese, 3-ethyl-2-oxoglutarate, and factor X substrate peptide fragment(39mer-4Ser) | | 分子名称: | (3~{R})-3-ethyl-2-oxidanylidene-pentanedioic acid, Aspartyl/asparaginyl beta-hydroxylase, Coagulation factor X, ... | | 著者 | Nakashima, Y, Brewitz, L, Schofield, C.J. | | 登録日 | 2020-05-29 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Synthesis of 2-oxoglutarate derivatives and their evaluation as cosubstrates and inhibitors of human aspartate/asparagine-beta-hydroxylase.
Chem Sci, 12, 2020
|
|
6Z6R
 
 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH) oxygenase and TPR domains in complex with manganese, N-oxalyl-alpha-methylalanine, and factor X substrate peptide fragment(39mer-4Ser) | | 分子名称: | Aspartyl/asparaginyl beta-hydroxylase, BROMIDE ION, Coagulation factor X, ... | | 著者 | Nakashima, Y, Brewitz, L, Schofield, C.J. | | 登録日 | 2020-05-29 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Synthesis of 2-oxoglutarate derivatives and their evaluation as cosubstrates and inhibitors of human aspartate/asparagine-beta-hydroxylase.
Chem Sci, 12, 2020
|
|
8G9T
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | 分子名称: | AcrIC9, Cas11, Cas5, ... | | 著者 | Hu, C, Nam, K.H, Ke, A. | | 登録日 | 2023-02-22 | | 公開日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8G9S
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | 分子名称: | AcrIC8, Cas11, Cas5, ... | | 著者 | Hu, C, Nam, K.H, Ke, A. | | 登録日 | 2023-02-22 | | 公開日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
7R72
 
 | | State E1 nucleolar 60S ribosome biogenesis intermediate - Spb4 local model | | 分子名称: | 25S rRNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, 5.8S rRNA, ... | | 著者 | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | 登録日 | 2021-06-24 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8G9U
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | 分子名称: | CRISPR-associated protein, Csd1 family, Csd2 family, ... | | 著者 | Hu, C, Nam, K.H, Ke, A. | | 登録日 | 2023-02-22 | | 公開日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
7R3F
 
 | | Monomeric PqsE mutant E187R | | 分子名称: | 2-aminobenzoylacetyl-CoA thioesterase, BENZOIC ACID, CACODYLATE ION, ... | | 著者 | Borgert, S.R, Schmelz, S, Blankenfeldt, W. | | 登録日 | 2022-02-07 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
