4FOS
 
 | | Crystal Structure of Shikimate Dehydrogenase (aroE) Q237A Mutant from Helicobacter pylori in Complex with Shikimate | | 分子名称: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, Shikimate dehydrogenase | | 著者 | Cheng, W.C, Chen, T.J, Wang, W.C. | | 登録日 | 2012-06-21 | | 公開日 | 2012-12-26 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 |
|
|
3ULD
 
 | |
6XBF
 
 | | Structure of NDM-1 in complex with macrocycle inhibitor NDM1i-1G | | 分子名称: | BlaNDM-4_1_JQ348841, ZINC ION, macrocycle inhibitor NDM1i-1G | | 著者 | Worrall, L.J, Sun, T, Mulligan, V.K, Strynadka, N.C.J. | | 登録日 | 2020-06-05 | | 公開日 | 2021-03-31 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Computationally designed peptide macrocycle inhibitors of New Delhi metallo-beta-lactamase 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3UVT
 
 | |
8F51
 
 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with mefloquine | | 分子名称: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | 登録日 | 2022-11-11 | | 公開日 | 2023-02-01 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
6XBE
 
 | | Structure of NDM-1 in complex with macrocycle inhibitor NDM1i-1F | | 分子名称: | BlaNDM-4_1_JQ348841, ZINC ION, macrocycle inhibitor NDM1i-1F | | 著者 | Worrall, L.J, Sun, T, Mulligan, V.K, Strynadka, N.C.J. | | 登録日 | 2020-06-05 | | 公開日 | 2021-03-31 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Computationally designed peptide macrocycle inhibitors of New Delhi metallo-beta-lactamase 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6XCI
 
 | | Structure of NDM-1 in complex with macrocycle inhibitor NDM1i-3D | | 分子名称: | ACETATE ION, BlaNDM-4_1_JQ348841, CADMIUM ION, ... | | 著者 | Worrall, L.J, Sun, T, Mulligan, V.K, Strynadka, N.C.J. | | 登録日 | 2020-06-08 | | 公開日 | 2021-03-31 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Computationally designed peptide macrocycle inhibitors of New Delhi metallo-beta-lactamase 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3UWI
 
 | | Bovine trypsin variant X(tripleGlu217Phe227) in complex with small molecule inhibitor | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | 著者 | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | 登録日 | 2011-12-02 | | 公開日 | 2012-12-05 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
4FPB
 
 | |
8EPJ
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 17 | | 分子名称: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-[(3-{[4-(morpholin-4-yl)-2-nitroanilino]methyl}phenyl)methyl]piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Karade, S.S, Mariuzza, R.A. | | 登録日 | 2022-10-05 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
6XG4
 
 | | X-ray structure of Escherichia coli dihydrofolate reductase L28R mutant in complex with trimethoprim | | 分子名称: | CHLORIDE ION, Dihydrofolate reductase, GLYCEROL, ... | | 著者 | Gaszek, I.K, Manna, M.S, Borek, D, Toprak, E. | | 登録日 | 2020-06-16 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A trimethoprim derivative impedes antibiotic resistance evolution.
Nat Commun, 12, 2021
|
|
4FQ8
 
 | | Crystal Structure of Shikimate Dehydrogenase (aroE) Y210A Mutant from Helicobacter pylori in Complex with Shikimate | | 分子名称: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, Shikimate dehydrogenase | | 著者 | Cheng, W.C, Chen, T.J, Wang, W.C. | | 登録日 | 2012-06-25 | | 公開日 | 2012-12-26 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 |
|
|
6XNO
 
 | | Crystal structure of E99A mutant of human CEACAM1 | | 分子名称: | Carcinoembryonic antigen-related cell adhesion molecule 1, MALONIC ACID, octyl beta-D-glucopyranoside | | 著者 | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Bonsor, D, Petsko, G.A, Kuchroo, V, Blumberg, R.S. | | 登録日 | 2020-07-03 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis of the dynamic human CEACAM1 monomer-dimer equilibrium.
Commun Biol, 4, 2021
|
|
8EGV
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 12 | | 分子名称: | (2R,3R,4R,5S)-1-{2-[4-(2-{[(5M)-3-chloro-5-(1,2,4-oxadiazol-3-yl)phenyl]amino}ethyl)phenyl]ethyl}-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Karade, S.S, Mariuzza, R.A. | | 登録日 | 2022-09-13 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
3UXG
 
 | | Crystal structure of RFXANK | | 分子名称: | DNA-binding protein RFXANK, Histone deacetylase 4, UNKNOWN ATOM OR ION | | 著者 | Tempel, W, Chao, X, Bian, C, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2011-12-05 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Sequence-Specific Recognition of a PxLPxI/L Motif by an Ankyrin Repeat Tumbler Lock.
Sci.Signal., 5, 2012
|
|
8EHP
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 13 | | 分子名称: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-[(4-{[4-(morpholin-4-yl)anilino]methyl}phenyl)methyl]piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Karade, S.S, Mariuzza, R.A. | | 登録日 | 2022-09-14 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8EID
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 14 | | 分子名称: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{[4-({[(5P)-3-(methanesulfonyl)-5-(pyridazin-3-yl)phenyl]amino}methyl)phenyl]methyl}piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Karade, S.S, Mariuzza, R.A. | | 登録日 | 2022-09-14 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
4ZIM
 
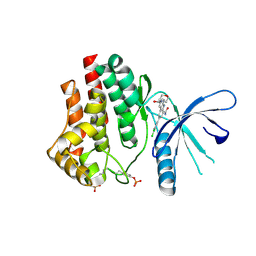 | |
3UN4
 
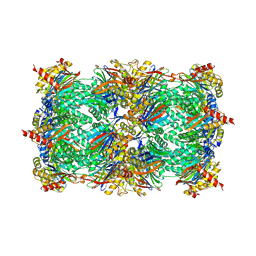 | | Yeast 20S proteasome in complex with PR-957 (morpholine) | | 分子名称: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, Proteasome component C1, Proteasome component C11, ... | | 著者 | Huber, E, Basler, M, Schwab, R, Heinemeyer, W, Kirk, C, Groettrup, M, Groll, M. | | 登録日 | 2011-11-15 | | 公開日 | 2012-02-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity.
Cell(Cambridge,Mass.), 148, 2012
|
|
6XG5
 
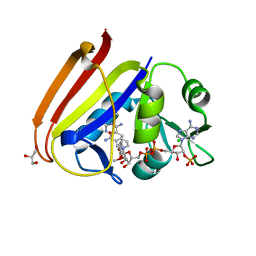 | | X-ray structure of Escherichia coli dihydrofolate reductase in complex with trimethoprim | | 分子名称: | CHLORIDE ION, Dihydrofolate reductase, GLYCEROL, ... | | 著者 | Gaszek, I.K, Manna, M.S, Borek, D, Toprak, E. | | 登録日 | 2020-06-16 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A trimethoprim derivative impedes antibiotic resistance evolution.
Nat Commun, 12, 2021
|
|
8ELE
 
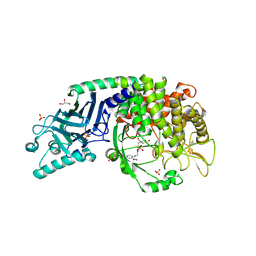 | | Co-crystal structure of Chaetomium glucosidase with compound 16 | | 分子名称: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{[4-({2-nitro-4-[(1R,5S)-3-oxa-8-azabicyclo[3.2.1]octan-8-yl]anilino}methyl)phenyl]methyl}piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | 著者 | Karade, S.S, Mariuzza, R.A. | | 登録日 | 2022-09-23 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
3UXL
 
 | |
4F9C
 
 | | Human CDC7 kinase in complex with DBF4 and XL413 | | 分子名称: | 8-chloro-2-[(2S)-pyrrolidin-2-yl][1]benzofuro[3,2-d]pyrimidin-4(3H)-one, Cell division cycle 7-related protein kinase, Protein DBF4 homolog A, ... | | 著者 | Hughes, S, Cherepanov, P. | | 登録日 | 2012-05-18 | | 公開日 | 2012-10-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Crystal structure of human CDC7 kinase in complex with its activator DBF4.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4FR2
 
 | | Alcohol dehydrogenase from Oenococcus oeni | | 分子名称: | 1,3-propanediol dehydrogenase, NICKEL (II) ION | | 著者 | Fodor, K, Skander, E, Wilmanns, M. | | 登録日 | 2012-06-26 | | 公開日 | 2013-06-05 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural and biochemical characterisation of a NAD(+)-dependent alcohol dehydrogenase from Oenococcus oeni as a new model molecule for industrial biotechnology applications.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
3UPE
 
 | | Bovine trypsin variant X(triplePhe227) in complex with small molecule inhibitor | | 分子名称: | 1-[(7-carbamimidoylnaphthalen-2-yl)methyl]-6-({1-[(1Z)-ethanimidoyl]piperidin-4-yl}oxy)-2-(propan-2-yl)-1H-indole-4-carboxylic acid, CALCIUM ION, Cationic trypsin, ... | | 著者 | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | 登録日 | 2011-11-18 | | 公開日 | 2012-11-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
