6M2V
 
 | | Crystal structure of UHRF1 SRA complexed with fully-mCHG DNA. | | 分子名称: | DNA (5'-D(*TP*CP*AP*CP*GP*(5CM)P*TP*GP*CP*GP*TP*GP*A)-3'), E3 ubiquitin-protein ligase UHRF1 | | 著者 | Abhishek, S, Nakarakanti, N.K, Deeksha, W, Rajakumara, E. | | 登録日 | 2020-03-01 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Mechanistic insights into recognition of symmetric methylated cytosines in CpG and non-CpG DNA by UHRF1 SRA.
Int.J.Biol.Macromol., 170, 2021
|
|
4XRH
 
 | |
5K5H
 
 | |
1DCT
 
 | | DNA (CYTOSINE-5) METHYLASE FROM HAEIII COVALENTLY BOUND TO DNA | | 分子名称: | CALCIUM ION, DNA (5'-D(*AP*CP*CP*AP*GP*CP*AP*GP*GP*(C49)P*CP*AP*CP*CP*AP*GP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*CP*TP*GP*GP*TP*GP*GP*(C5M)P*CP*TP*GP*CP*TP*GP*G)-3'), ... | | 著者 | Reinisch, K.M, Chen, L, Verdine, G.L, Lipscomb, W.N. | | 登録日 | 1995-05-17 | | 公開日 | 1995-09-15 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The crystal structure of HaeIII methyltransferase convalently complexed to DNA: an extrahelical cytosine and rearranged base pairing.
Cell(Cambridge,Mass.), 82, 1995
|
|
1DNE
 
 | | MOLECULAR STRUCTURE OF THE NETROPSIN-D(CGCGATATCGCG) COMPLEX: DNA CONFORMATION IN AN ALTERNATING AT SEGMENT; CONFORMATION 2 | | 分子名称: | DNA (5'-D(*CP*GP*CP*GP*AP*TP*AP*TP*CP*GP*CP*G)-3'), NETROPSIN | | 著者 | Coll, M, Aymami, J, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | 登録日 | 1988-09-14 | | 公開日 | 1989-01-09 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular structure of the netropsin-d(CGCGATATCGCG) complex: DNA conformation in an alternating AT segment.
Biochemistry, 28, 1989
|
|
5GAT
 
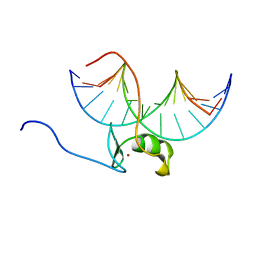 | | SOLUTION NMR STRUCTURE OF THE WILD TYPE DNA BINDING DOMAIN OF AREA COMPLEXED TO A 13BP DNA CONTAINING A CGATA SITE, 35 STRUCTURES | | 分子名称: | DNA (5'-D(*CP*AP*GP*CP*GP*AP*TP*AP*GP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*CP*TP*AP*TP*CP*GP*CP*TP*G)-3'), NITROGEN REGULATORY PROTEIN AREA, ... | | 著者 | Clore, G.M, Starich, M, Wikstrom, M, Gronenborn, A.M. | | 登録日 | 1997-11-07 | | 公開日 | 1998-01-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of a fungal AREA protein-DNA complex: an alternative binding mode for the basic carboxyl tail of GATA factors.
J.Mol.Biol., 277, 1998
|
|
5KKQ
 
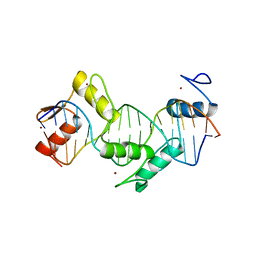 | | Homo sapiens CCCTC-binding factor (CTCF) ZnF3-7 and DNA complex structure | | 分子名称: | DNA (5'-D(*GP*CP*CP*AP*GP*CP*AP*GP*GP*GP*GP*GP*CP*GP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*CP*GP*CP*CP*CP*CP*CP*TP*GP*CP*TP*GP*GP*C)-3'), Transcriptional repressor CTCF, ... | | 著者 | Hashimoto, H, Wang, D, Cheng, X. | | 登録日 | 2016-06-22 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.744 Å) | | 主引用文献 | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
8VX4
 
 | |
8VX6
 
 | |
5DB8
 
 | |
7UXD
 
 | |
5DB6
 
 | |
5DBC
 
 | |
5DB7
 
 | |
7UI4
 
 | |
8E86
 
 | |
8E8B
 
 | |
8E87
 
 | |
8E8J
 
 | |
8E8G
 
 | |
8E3E
 
 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing CAST sequence (#10) | | 分子名称: | DNA (5'-D(*CP*TP*TP*TP*GP*GP*GP*GP*AP*GP*GP*GP*GP*TP*CP*TP*TP*TP*TP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*AP*AP*AP*GP*AP*CP*CP*CP*CP*TP*CP*CP*CP*CP*AP*AP*A)-3'), ZINC ION, ... | | 著者 | Horton, J.R, Ren, R, Cheng, X. | | 登録日 | 2022-08-17 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
8E8D
 
 | |
8E85
 
 | |
8E8F
 
 | |
8E8C
 
 | |
