1B99
 
 | | 3'-FLUORO-URIDINE DIPHOSPHATE BINDING TO NUCLEOSIDE DIPHOSPHATE KINASE | | 分子名称: | 2',3'-DIDEOXY-3'-FLUORO-URIDIDINE-5'-DIPHOSPHATE, PROTEIN (NUCLEOSIDE DIPHOSPHATE KINASE), PYROPHOSPHATE 2- | | 著者 | Janin, J, Xu, Y. | | 登録日 | 1999-02-22 | | 公開日 | 1999-06-28 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Catalytic mechanism of nucleoside diphosphate kinase investigated using nucleotide analogues, viscosity effects, and X-ray crystallography.
Biochemistry, 38, 1999
|
|
7ZL8
 
 | | NME1 in complex with succinyl-CoA | | 分子名称: | Nucleoside diphosphate kinase A, SUCCINYL-COENZYME A | | 著者 | Garcia-Saez, I, Iuso, D, Khochbin, S, Petosa, C. | | 登録日 | 2022-04-14 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Nucleoside diphosphate kinases 1 and 2 regulate a protective liver response to a high-fat diet.
Sci Adv, 9, 2023
|
|
1BHN
 
 | | NUCLEOSIDE DIPHOSPHATE KINASE ISOFORM A FROM BOVINE RETINA | | 分子名称: | GUANOSINE-3',5'-MONOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, NUCLEOSIDE DIPHOSPHATE TRANSFERASE | | 著者 | Ladner, J.E, Abdulaev, N.G, Kakuev, D.L, Karaschuk, G.N, Tordova, M, Eisenstein, E, Fujiwara, J.H, Ridge, K.D, Gilliland, G.L. | | 登録日 | 1998-06-10 | | 公開日 | 1999-02-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The three-dimensional structures of two isoforms of nucleoside diphosphate kinase from bovine retina.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
4OQP
 
 | |
4OQQ
 
 | |
3Q86
 
 | |
1VHB
 
 | | BACTERIAL DIMERIC HEMOGLOBIN FROM VITREOSCILLA STERCORARIA | | 分子名称: | HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Tarricone, C, Galizzi, A, Coda, A, Ascenzi, P, Bolognesi, M. | | 登録日 | 1997-02-19 | | 公開日 | 1998-02-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Unusual structure of the oxygen-binding site in the dimeric bacterial hemoglobin from Vitreoscilla sp.
Structure, 5, 1997
|
|
3Q8Y
 
 | |
3Q8V
 
 | |
1EHW
 
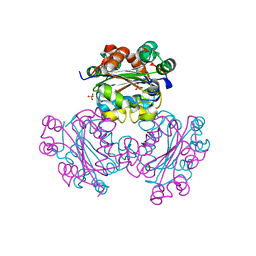 | | HUMAN NUCLEOSIDE DIPHOSPHATE KINASE 4 | | 分子名称: | NUCLEOSIDE DIPHOSPHATE KINASE, SULFATE ION | | 著者 | Milon, L, Meyer, P, Chiadmi, M, Munier, A, Johansson, M, Karlsson, A, Lascu, I, Capeau, J, Janin, J, Lacombe, M.-L. | | 登録日 | 2000-02-23 | | 公開日 | 2000-05-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The human nm23-H4 gene product is a mitochondrial nucleoside diphosphate kinase.
J.Biol.Chem., 275, 2000
|
|
3Q89
 
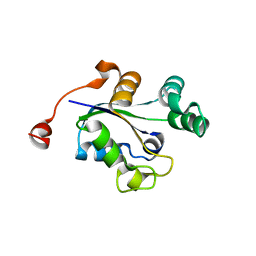 | |
3Q8U
 
 | |
7ABI
 
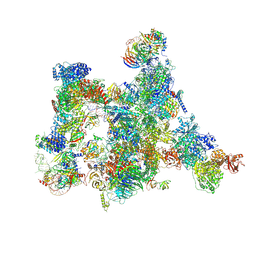 | | Human pre-Bact-2 spliceosome | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Beta-catenin-like protein 1, ... | | 著者 | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | 登録日 | 2020-09-07 | | 公開日 | 2021-02-10 | | 最終更新日 | 2025-04-09 | | 実験手法 | ELECTRON MICROSCOPY (8 Å) | | 主引用文献 | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
7ABG
 
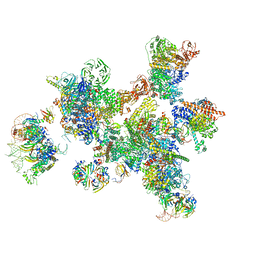 | | Human pre-Bact-1 spliceosome | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Cell division cycle 5-like protein, ... | | 著者 | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | 登録日 | 2020-09-07 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (7.8 Å) | | 主引用文献 | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
3NGS
 
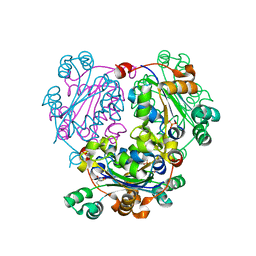 | | Structure of Leishmania nucleoside diphosphate kinase b with ordered nucleotide-binding loop | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Nucleoside diphosphate kinase, PHOSPHATE ION | | 著者 | Trindade, D.M, Sousa, T.A.C.B, Tonoli, C.C.C, Santos, C.R, Arni, R.K, Ward, R.J, Oliveira, A.H.C, Murakami, M.T. | | 登録日 | 2010-06-13 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular adaptability of nucleoside diphosphate kinase b from trypanosomatid parasites: stability, oligomerization and structural determinants of nucleotide binding.
Mol Biosyst, 7, 2011
|
|
7AAV
 
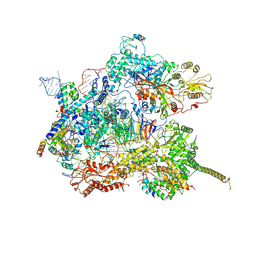 | | Human pre-Bact-2 spliceosome core structure | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, D-chiro inositol hexakisphosphate, ... | | 著者 | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | 登録日 | 2020-09-04 | | 公開日 | 2020-12-09 | | 最終更新日 | 2025-04-09 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
7ABF
 
 | | Human pre-Bact-1 spliceosome core structure | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | 登録日 | 2020-09-07 | | 公開日 | 2020-12-09 | | 最終更新日 | 2025-04-09 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
1H1H
 
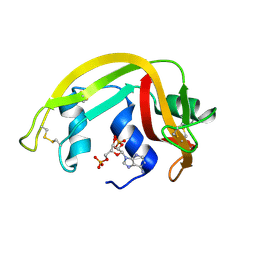 | | Crystal Structure of Eosinophil Cationic Protein in Complex with 2',5'-ADP at 2.0 A resolution Reveals the Details of the Ribonucleolytic Active site | | 分子名称: | ADENOSINE-2'-5'-DIPHOSPHATE, EOSINOPHIL CATIONIC PROTEIN | | 著者 | Mohan, C.G, Boix, E, Evans, H.R, Nikolovski, Z, Nogues, M.V, Cuchillo, C.M, Acharya, K.R. | | 登録日 | 2002-07-15 | | 公開日 | 2002-10-03 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Crystal Structure of Eosinophil Cationic Protein in Complex with 2'5'-Adp at 2.0 A Resolution Reveals the Details of the Ribonucleolytic Active Site
Biochemistry, 41, 2002
|
|
3VHB
 
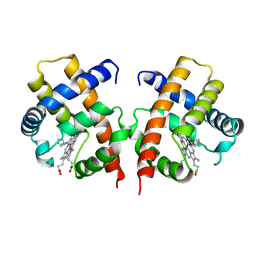 | | IMIDAZOLE ADDUCT OF THE BACTERIAL HEMOGLOBIN FROM VITREOSCILLA SP. | | 分子名称: | IMIDAZOLE, PROTEIN (HEMOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Bolognesi, M, Boffi, A, Coletta, M, Mozzarelli, A, Pesce, A, Tarricone, C, Ascenzi, P. | | 登録日 | 1999-03-17 | | 公開日 | 1999-08-18 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Anticooperative ligand binding properties of recombinant ferric Vitreoscilla homodimeric hemoglobin: a thermodynamic, kinetic and X-ray crystallographic study.
J.Mol.Biol., 291, 1999
|
|
1H8U
 
 | | Crystal Structure of the Eosinophil Major Basic Protein at 1.8A: An Atypical Lectin with a Paradigm Shift in Specificity | | 分子名称: | EOSINOPHIL GRANULE MAJOR BASIC PROTEIN 1, GLYCEROL, SULFATE ION | | 著者 | Swaminathan, G.J, Weaver, A.J, Loegering, D.A, Checkel, J.L, Leonidas, D.D, Gleich, G.J, Acharya, K.R. | | 登録日 | 2001-02-15 | | 公開日 | 2001-07-17 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of the Eosinophil Major Basic Protein at 1.8A. An Atypical Lectin with a Paradigm Shift in Specificity
J.Biol.Chem., 276, 2001
|
|
1QGV
 
 | | HUMAN SPLICEOSOMAL PROTEIN U5-15KD | | 分子名称: | SPLICEOSOMAL PROTEIN U5-15KD | | 著者 | Reuter, K, Nottrott, S, Fabrizio, P, Luehrmann, R, Ficner, R. | | 登録日 | 1999-05-06 | | 公開日 | 1999-12-12 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Identification, characterization and crystal structure analysis of the human spliceosomal U5 snRNP-specific 15 kD protein.
J.Mol.Biol., 294, 1999
|
|
3TM9
 
 | | Y29A mutant of Vitreoscilla stercoraria hemoglobin | | 分子名称: | 1,2-ETHANEDIOL, Bacterial hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Ratakonda, S, Anand, A, Dikshit, K, Stark, B.C, Howard, A.J. | | 登録日 | 2011-08-31 | | 公開日 | 2014-04-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Crystallographic structure determination of B10 mutants of Vitreoscilla hemoglobin: role of Tyr29 (B10) in the structure of the ligand-binding site.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3TM3
 
 | | Wild-type hemoglobin from Vitreoscilla stercoraria | | 分子名称: | Hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Ratakonda, S, Anand, A, Dikshit, K, Stark, B.C, Howard, A.J. | | 登録日 | 2011-08-31 | | 公開日 | 2014-04-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystallographic structure determination of B10 mutants of Vitreoscilla hemoglobin: role of Tyr29 (B10) in the structure of the ligand-binding site.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3TLD
 
 | | Crystal Structure of Y29F mutant of Vitreoscilla hemoglobin | | 分子名称: | Bacterial hemoglobin, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Ratakonda, S, Anand, A, Dikshit, K, Stark, B.C, Howard, A.J. | | 登録日 | 2011-08-29 | | 公開日 | 2014-04-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.896 Å) | | 主引用文献 | Crystallographic structure determination of B10 mutants of Vitreoscilla hemoglobin: role of Tyr29 (B10) in the structure of the ligand-binding site.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
1ZUX
 
 | |
